electroChem Package Documentation¶
This page contains the electroChem Package documentation.
The gapFillMesh Module¶
The GapFillMesh object glues 3 meshes together to form a composite mesh. The first mesh is a Grid2D object that is fine and deals with the area around the trench or via. The second mesh is a GmshImporter2D object that forms a transition mesh from a fine to a course region. The third mesh is another Grid2D object that forms the boundary layer. This region consists of very large elements and is only used for the diffusion in the boundary layer.
- class fipy.models.levelSet.electroChem.gapFillMesh.GapFillMesh(cellSize=None, desiredDomainWidth=None, desiredDomainHeight=None, desiredFineRegionHeight=None, transitionRegionHeight=None)¶
Bases: fipy.meshes.numMesh.mesh2D.Mesh2D
The following test case tests for diffusion across the domain.
>>> domainHeight = 5. >>> mesh = GapFillMesh(transitionRegionHeight = 2., ... cellSize = 0.1, ... desiredFineRegionHeight = 1., ... desiredDomainHeight = domainHeight, ... desiredDomainWidth = 1.)
>>> import fipy.tools.dump as dump >>> (f, filename) = dump.write(mesh) >>> mesh = dump.read(filename, f) >>> mesh.getNumberOfCells() - len(mesh.getCellIDsAboveFineRegion()) 90
>>> from fipy.variables.cellVariable import CellVariable >>> var = CellVariable(mesh = mesh)
>>> from fipy.terms.diffusionTerm import DiffusionTerm >>> eq = DiffusionTerm()
>>> from fipy.boundaryConditions.fixedValue import FixedValue >>> eq.solve(var, boundaryConditions = (FixedValue(mesh.getFacesBottom(), 0.), ... FixedValue(mesh.getFacesTop(), domainHeight)))
Evaluate the result:
>>> centers = mesh.getCellCenters()[1].copy() ## the copy makes the array contiguous for inlining >>> localErrors = (centers - var)**2 / centers**2 >>> globalError = numerix.sqrt(numerix.sum(localErrors) / mesh.getNumberOfCells()) >>> argmax = numerix.argmax(localErrors) >>> print numerix.sqrt(localErrors[argmax]) < 0.1 1 >>> print globalError < 0.05 1
Arguments:
cellSize - The cell size in the fine grid around the trench.
desiredDomainWidth - The desired domain width.
desiredDomainHeight - The total desired height of the domain.
desiredFineRegionHeight - The desired height of the in the fine region around the trench.
transitionRegionHeight - The height of the transition region.
- buildTransitionMesh(nx, height, cellSize)¶
- getCellIDsAboveFineRegion()¶
- getFineMesh()¶
- class fipy.models.levelSet.electroChem.gapFillMesh.TrenchMesh(trenchDepth=None, trenchSpacing=None, boundaryLayerDepth=None, cellSize=None, aspectRatio=None, angle=0.0, bowWidth=0.0, overBumpRadius=0.0, overBumpWidth=0.0)¶
Bases: fipy.models.levelSet.electroChem.gapFillMesh.GapFillMesh
The trench mesh takes the parameters generally used to define a trench region and recasts then for the general GapFillMesh.
The following test case tests for diffusion across the domain.
>>> cellSize = 0.05e-6 >>> trenchDepth = 0.5e-6 >>> boundaryLayerDepth = 50e-6 >>> domainHeight = 10 * cellSize + trenchDepth + boundaryLayerDepth
>>> mesh = TrenchMesh(trenchSpacing = 1e-6, ... cellSize = cellSize, ... trenchDepth = trenchDepth, ... boundaryLayerDepth = boundaryLayerDepth, ... aspectRatio = 1.)
>>> import fipy.tools.dump as dump >>> (f, filename) = dump.write(mesh) >>> mesh = dump.read(filename, f) >>> mesh.getNumberOfCells() - len(numerix.nonzero(mesh.getElectrolyteMask())[0]) 150
>>> from fipy.variables.cellVariable import CellVariable >>> var = CellVariable(mesh = mesh, value = 0.)
>>> from fipy.terms.diffusionTerm import DiffusionTerm >>> eq = DiffusionTerm()
>>> from fipy.boundaryConditions.fixedValue import FixedValue
>>> eq.solve(var, boundaryConditions = (FixedValue(mesh.getFacesBottom(), 0.), ... FixedValue(mesh.getFacesTop(), domainHeight)))
Evaluate the result:
>>> centers = mesh.getCellCenters()[1].copy() ## ensure contiguous array for inlining >>> localErrors = (centers - var)**2 / centers**2 >>> globalError = numerix.sqrt(numerix.sum(localErrors) / mesh.getNumberOfCells()) >>> argmax = numerix.argmax(localErrors) >>> print numerix.sqrt(localErrors[argmax]) < 0.051 1 >>> print globalError < 0.02 1
trenchDepth - Depth of the trench.
trenchSpacing - The distance between the trenches.
boundaryLayerDepth - The depth of the hydrodynamic boundary layer.
cellSize - The cell Size.
aspectRatio - trenchDepth / trenchWidth
angle - The angle for the taper of the trench.
bowWidth - The maximum displacement for any bow in the trench shape.
overBumpWidth - The width of the over bump.
overBumpRadius - The radius of the over bump.
- getBottomFaces()¶
Included to not break the interface
- getElectrolyteMask()¶
- getTopFaces()¶
Included to not break the interface
The metalIonDiffusionEquation Module¶
- fipy.models.levelSet.electroChem.metalIonDiffusionEquation.buildMetalIonDiffusionEquation(ionVar=None, distanceVar=None, depositionRate=1, transientCoeff=1, diffusionCoeff=1, metalIonMolarVolume=1)¶
The MetalIonDiffusionEquation solves the diffusion of the metal species with a source term at the electrolyte interface. The governing equation is given by,
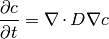
where,
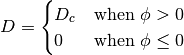
The velocity of the interface generally has a linear dependence on ion concentration. The following boundary condition applies at the zero level set,
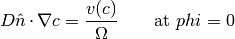
where
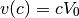
The test case below is for a 1D steady state problem. The solution is given by:
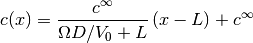
This is the test case,
>>> from fipy.meshes.grid1D import Grid1D >>> nx = 11 >>> dx = 1. >>> from fipy.tools import serial >>> mesh = Grid1D(nx = nx, dx = dx, communicator=serial) >>> x, = mesh.getCellCenters() >>> from fipy.variables.cellVariable import CellVariable >>> ionVar = CellVariable(mesh = mesh, value = 1.) >>> from fipy.models.levelSet.distanceFunction.distanceVariable \ ... import DistanceVariable >>> disVar = DistanceVariable(mesh = mesh, ... value = (x - 0.5) - 0.99, ... hasOld = 1)
>>> v = 1. >>> diffusion = 1. >>> omega = 1. >>> cinf = 1. >>> from fipy.boundaryConditions.fixedValue import FixedValue >>> eqn = buildMetalIonDiffusionEquation(ionVar = ionVar, ... distanceVar = disVar, ... depositionRate = v * ionVar, ... diffusionCoeff = diffusion, ... metalIonMolarVolume = omega) >>> bc = (FixedValue(mesh.getFacesRight(), cinf),) >>> for i in range(10): ... eqn.solve(ionVar, dt = 1000, boundaryConditions = bc) >>> L = (nx - 1) * dx - dx / 2 >>> gradient = cinf / (omega * diffusion / v + L) >>> answer = gradient * (x - L - dx * 3 / 2) + cinf >>> answer[x < dx] = 1 >>> print ionVar.allclose(answer) 1
Parameters : - ionVar: The metal ion concentration variable.
- distanceVar: A DistanceVariable object.
- depositionRate: A float or a CellVariable representing the interface deposition rate.
- transientCoeff: The transient coefficient.
- diffusionCoeff: The diffusion coefficient
- metalIonMolarVolume: Molar volume of the metal ions.

 FiPy
FiPy