distanceFunction Package Documentation¶
This page contains the distanceFunction Package documentation.
The distanceVariable Module¶
- class fipy.models.levelSet.distanceFunction.distanceVariable.DistanceVariable(mesh, name='', value=0.0, unit=None, hasOld=0, narrowBandWidth=10000000000.0)¶
Bases: fipy.variables.cellVariable.CellVariable
A DistanceVariable object calculates
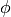 so it satisfies,
so it satisfies,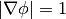
using the fast marching method with an initial condition defined by the zero level set.
Currently the solution is first order, This suffices for initial conditions with straight edges (e.g. trenches in electrodeposition). The method should work for unstructured 2D grids but testing on unstructured grids is untested thus far. This is a 2D implementation as it stands. Extending to 3D should be relatively simple.
Here we will define a few test cases. Firstly a 1D test case
>>> from fipy.meshes.grid1D import Grid1D >>> from fipy.tools import serial >>> mesh = Grid1D(dx = .5, nx = 8, communicator=serial) >>> from distanceVariable import DistanceVariable >>> var = DistanceVariable(mesh = mesh, value = (-1, -1, -1, -1, 1, 1, 1, 1)) >>> var.calcDistanceFunction() >>> answer = (-1.75, -1.25, -.75, -0.25, 0.25, 0.75, 1.25, 1.75) >>> print var.allclose(answer) 1
A 1D test case with very small dimensions.
>>> dx = 1e-10 >>> mesh = Grid1D(dx = dx, nx = 8, communicator=serial) >>> var = DistanceVariable(mesh = mesh, value = (-1, -1, -1, -1, 1, 1, 1, 1)) >>> var.calcDistanceFunction() >>> answer = numerix.arange(8) * dx - 3.5 * dx >>> print var.allclose(answer) 1
A 2D test case to test _calcTrialValue for a pathological case.
>>> dx = 1. >>> dy = 2. >>> from fipy.meshes.grid2D import Grid2D >>> mesh = Grid2D(dx = dx, dy = dy, nx = 2, ny = 3) >>> var = DistanceVariable(mesh = mesh, value = (-1, 1, 1, 1, -1, 1))
>>> var.calcDistanceFunction() >>> vbl = -dx * dy / numerix.sqrt(dx**2 + dy**2) / 2. >>> vbr = dx / 2 >>> vml = dy / 2. >>> crossProd = dx * dy >>> dsq = dx**2 + dy**2 >>> top = vbr * dx**2 + vml * dy**2 >>> sqrt = crossProd**2 *(dsq - (vbr - vml)**2) >>> sqrt = numerix.sqrt(max(sqrt, 0)) >>> vmr = (top + sqrt) / dsq >>> answer = (vbl, vbr, vml, vmr, vbl, vbr) >>> print var.allclose(answer) 1
The extendVariable method solves the following equation for a given extensionVariable.
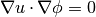
using the fast marching method with an initial condition defined at the zero level set. Essentially the equation solves a fake distance function to march out the velocity from the interface.
>>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(dx = 1., dy = 1., nx = 2, ny = 2) >>> var = DistanceVariable(mesh = mesh, value = (-1, 1, 1, 1)) >>> var.calcDistanceFunction() >>> extensionVar = CellVariable(mesh = mesh, value = (-1, .5, 2, -1)) >>> tmp = 1 / numerix.sqrt(2) >>> print var.allclose((-tmp / 2, 0.5, 0.5, 0.5 + tmp)) 1 >>> var.extendVariable(extensionVar) >>> print extensionVar.allclose((1.25, .5, 2, 1.25)) 1 >>> mesh = Grid2D(dx = 1., dy = 1., nx = 3, ny = 3) >>> var = DistanceVariable(mesh = mesh, value = (-1, 1, 1, ... 1, 1, 1, ... 1, 1, 1)) >>> var.calcDistanceFunction() >>> extensionVar = CellVariable(mesh = mesh, value = (-1, .5, -1, ... 2, -1, -1, ... -1, -1, -1))
>>> v1 = 0.5 + tmp >>> v2 = 1.5 >>> tmp1 = (v1 + v2) / 2 + numerix.sqrt(2. - (v1 - v2)**2) / 2 >>> tmp2 = tmp1 + 1 / numerix.sqrt(2) >>> print var.allclose((-tmp / 2, 0.5, 1.5, 0.5, 0.5 + tmp, ... tmp1, 1.5, tmp1, tmp2)) 1 >>> answer = (1.25, .5, .5, 2, 1.25, 0.9544, 2, 1.5456, 1.25) >>> var.extendVariable(extensionVar) >>> print extensionVar.allclose(answer, rtol = 1e-4) 1
Test case for a bug that occurs when initializing the distance variable at the interface. Currently it is assumed that adjacent cells that are opposite sign neighbors have perpendicular normal vectors. In fact the two closest cells could have opposite normals.
>>> mesh = Grid1D(dx = 1., nx = 3) >>> var = DistanceVariable(mesh = mesh, value = (-1, 1, -1)) >>> var.calcDistanceFunction() >>> print var.allclose((-0.5, 0.5, -0.5)) 1
For future reference, the minimum distance for the interface cells can be calculated with the following functions. The trial cell values will also be calculated with these functions. In essence it is not difficult to calculate the level set distance function on an unstructured 3D grid. However a lot of testing will be required. The minimum distance functions will take the following form.
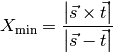
and in 3D,
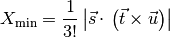
where the vectors
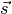 ,
, 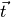 and
and 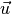 represent the
vectors from the cell of interest to the neighboring cell.
represent the
vectors from the cell of interest to the neighboring cell.Creates a distanceVariable object.
Parameters : - mesh: The mesh that defines the geometry of this variable.
- name: The name of the variable.
- value: The initial value.
- unit: the physical units of the variable
- hasOld: Whether the variable maintains an old value.
- narrowBandWidth: The width of the region about the zero level set within which the distance function is evaluated.
- calcDistanceFunction(narrowBandWidth=None, deleteIslands=False)¶
Calculates the distanceVariable as a distance function.
Parameters : - narrowBandWidth: The width of the region about the zero level set within which the distance function is evaluated.
- deleteIslands: Sets the temporary level set value to zero in isolated cells.
- extendVariable(extensionVariable, deleteIslands=False)¶
Takes a cellVariable and extends the variable from the zero to the region encapuslated by the narrowBandWidth.
Parameters : - extensionVariable: The variable to extend from the zero level set.
- deleteIslands: Sets the temporary level set value to zero in isolated cells.
- getCellInterfaceAreas()¶
Returns the length of the interface that crosses the cell
A simple 1D test:
>>> from fipy.meshes.grid1D import Grid1D >>> mesh = Grid1D(dx = 1., nx = 4) >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (-1.5, -0.5, 0.5, 1.5)) >>> answer = CellVariable(mesh=mesh, value=(0, 0., 1., 0)) >>> print numerix.allclose(distanceVariable.getCellInterfaceAreas(), ... answer) True
A 2D test case:
>>> from fipy.meshes.grid2D import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(dx = 1., dy = 1., nx = 3, ny = 3) >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (1.5, 0.5, 1.5, ... 0.5,-0.5, 0.5, ... 1.5, 0.5, 1.5)) >>> answer = CellVariable(mesh=mesh, ... value=(0, 1, 0, 1, 0, 1, 0, 1, 0)) >>> print numerix.allclose(distanceVariable.getCellInterfaceAreas(), answer) True
Another 2D test case:
>>> mesh = Grid2D(dx = .5, dy = .5, nx = 2, ny = 2) >>> from fipy.variables.cellVariable import CellVariable >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (-0.5, 0.5, 0.5, 1.5)) >>> answer = CellVariable(mesh=mesh, ... value=(0, numerix.sqrt(2) / 4, numerix.sqrt(2) / 4, 0)) >>> print numerix.allclose(distanceVariable.getCellInterfaceAreas(), ... answer) True
Test to check that the circumfrence of a circle is, in fact,
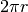 .
.>>> mesh = Grid2D(dx = 0.05, dy = 0.05, nx = 20, ny = 20) >>> r = 0.25 >>> x, y = mesh.getCellCenters() >>> rad = numerix.sqrt((x - .5)**2 + (y - .5)**2) - r >>> distanceVariable = DistanceVariable(mesh = mesh, value = rad) >>> print distanceVariable.getCellInterfaceAreas().sum() 1.57984690073

 FiPy
FiPy