surfactant Package Documentation¶
This page contains the surfactant Package documentation.
The adsorbingSurfactantEquation Module¶
- class fipy.models.levelSet.surfactant.adsorbingSurfactantEquation.AdsorbingSurfactantEquation(surfactantVar=None, distanceVar=None, bulkVar=None, rateConstant=None, otherVar=None, otherBulkVar=None, otherRateConstant=None, consumptionCoeff=None)¶
Bases: fipy.models.levelSet.surfactant.surfactantEquation.SurfactantEquation
The AdsorbingSurfactantEquation object solves the SurfactantEquation but with an adsorbing species from some bulk value. The equation that describes the surfactant adsorbing is given by,
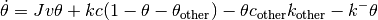
where
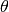 ,
, 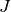 ,
,  ,
, 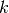 ,
,  ,
,
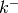 and
and 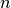 represent the surfactant coverage, the curvature,
the interface normal velocity, the adsorption rate, the concentration in the
bulk at the interface, the consumption rate and an exponent of consumption,
respectively. The
represent the surfactant coverage, the curvature,
the interface normal velocity, the adsorption rate, the concentration in the
bulk at the interface, the consumption rate and an exponent of consumption,
respectively. The 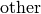 subscript refers to another
surfactant with greater surface affinity.
subscript refers to another
surfactant with greater surface affinity.The terms on the RHS of the above equation represent conservation of surfactant on a non-uniform surface, Langmuir adsorption, removal of surfactant due to adsorption of the other surfactant onto non-vacant sites and consumption of the surfactant respectively. The adsorption term is added to the source by setting :math:` S_c = k c (1 - theta_{text{other}})` and
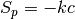 . The other terms are added to the source in a similar
way.
. The other terms are added to the source in a similar
way.The following is a test case:
>>> from fipy.models.levelSet.distanceFunction.distanceVariable \ ... import DistanceVariable >>> from fipy.models.levelSet.surfactant.surfactantVariable \ ... import SurfactantVariable >>> from fipy.meshes.grid2D import Grid2D >>> dx = .5 >>> dy = 2.3 >>> dt = 0.25 >>> k = 0.56 >>> initialValue = 0.1 >>> c = 0.2
>>> from fipy.meshes.grid2D import Grid2D >>> mesh = Grid2D(dx = dx, dy = dy, nx = 5, ny = 1) >>> distanceVar = DistanceVariable(mesh = mesh, ... value = (-dx*3/2, -dx/2, dx/2, ... 3*dx/2, 5*dx/2), ... hasOld = 1) >>> surfactantVar = SurfactantVariable(value = (0, 0, initialValue, 0 ,0), ... distanceVar = distanceVar) >>> bulkVar = CellVariable(mesh = mesh, value = (c , c, c, c, c)) >>> eqn = AdsorbingSurfactantEquation(surfactantVar = surfactantVar, ... distanceVar = distanceVar, ... bulkVar = bulkVar, ... rateConstant = k) >>> eqn.solve(surfactantVar, dt = dt) >>> answer = (initialValue + dt * k * c) / (1 + dt * k * c) >>> print numerix.allclose(surfactantVar.getInterfaceVar(), ... numerix.array((0, 0, answer, 0, 0))) 1
The following test case is for two surfactant variables. One has more surface affinity than the other.
>>> from fipy.models.levelSet.distanceFunction.distanceVariable \ ... import DistanceVariable >>> from fipy.models.levelSet.surfactant.surfactantVariable \ ... import SurfactantVariable >>> from fipy.meshes.grid2D import Grid2D >>> dx = 0.5 >>> dy = 2.73 >>> dt = 0.001 >>> k0 = 1. >>> k1 = 10. >>> theta0 = 0. >>> theta1 = 0. >>> c0 = 1. >>> c1 = 1. >>> totalSteps = 100 >>> mesh = Grid2D(dx = dx, dy = dy, nx = 5, ny = 1) >>> distanceVar = DistanceVariable(mesh = mesh, ... value = dx * (numerix.arange(5) - 1.5), ... hasOld = 1) >>> var0 = SurfactantVariable(value = (0, 0, theta0, 0 ,0), ... distanceVar = distanceVar) >>> var1 = SurfactantVariable(value = (0, 0, theta1, 0 ,0), ... distanceVar = distanceVar) >>> bulkVar0 = CellVariable(mesh = mesh, value = (c0, c0, c0, c0, c0)) >>> bulkVar1 = CellVariable(mesh = mesh, value = (c1, c1, c1, c1, c1))
>>> eqn0 = AdsorbingSurfactantEquation(surfactantVar = var0, ... distanceVar = distanceVar, ... bulkVar = bulkVar0, ... rateConstant = k0)
>>> eqn1 = AdsorbingSurfactantEquation(surfactantVar = var1, ... distanceVar = distanceVar, ... bulkVar = bulkVar1, ... rateConstant = k1, ... otherVar = var0, ... otherBulkVar = bulkVar0, ... otherRateConstant = k0)
>>> for step in range(totalSteps): ... eqn0.solve(var0, dt = dt) ... eqn1.solve(var1, dt = dt) >>> answer0 = 1 - numerix.exp(-k0 * c0 * dt * totalSteps) >>> answer1 = (1 - numerix.exp(-k1 * c1 * dt * totalSteps)) * (1 - answer0) >>> print numerix.allclose(var0.getInterfaceVar(), ... numerix.array((0, 0, answer0, 0, 0)), rtol = 1e-2) 1 >>> print numerix.allclose(var1.getInterfaceVar(), ... numerix.array((0, 0, answer1, 0, 0)), rtol = 1e-2) 1 >>> dt = 0.1 >>> for step in range(10): ... eqn0.solve(var0, dt = dt) ... eqn1.solve(var1, dt = dt)
>>> x, y = mesh.getCellCenters() >>> check = var0.getInterfaceVar() + var1.getInterfaceVar() >>> answer = CellVariable(mesh=mesh, value=check) >>> answer[x==1.25] = 1. >>> print check.allequal(answer) True
The following test case is to fix a bug where setting the adosrbtion coefficient to zero leads to the solver not converging and an eventual failure.
>>> var0 = SurfactantVariable(value = (0, 0, theta0, 0 ,0), ... distanceVar = distanceVar) >>> bulkVar0 = CellVariable(mesh = mesh, value = (c0, c0, c0, c0, c0))
>>> eqn0 = AdsorbingSurfactantEquation(surfactantVar = var0, ... distanceVar = distanceVar, ... bulkVar = bulkVar0, ... rateConstant = 0)
>>> eqn0.solve(var0, dt = dt) >>> eqn0.solve(var0, dt = dt) >>> answer = CellVariable(mesh=mesh, value=var0.getInterfaceVar()) >>> answer[x==1.25] = 0. >>> print var0.getInterfaceVar().allclose(answer) True
The following test case is to fix a bug that allows the accelerator to become negative.
>>> nx = 5 >>> ny = 5 >>> dx = 1. >>> dy = 1. >>> mesh = Grid2D(dx=dx, dy=dy, nx = nx, ny = ny) >>> x, y = mesh.getCellCenters() >>> disVar = DistanceVariable(mesh=mesh, value=1., hasOld=True) >>> disVar[y < dy] = -1 >>> disVar[x < dx] = -1 >>> disVar.calcDistanceFunction()
>>> levVar = SurfactantVariable(value = 0.5, distanceVar = disVar) >>> accVar = SurfactantVariable(value = 0.5, distanceVar = disVar)
>>> levEq = AdsorbingSurfactantEquation(levVar, ... distanceVar = disVar, ... bulkVar = 0, ... rateConstant = 0)
>>> accEq = AdsorbingSurfactantEquation(accVar, ... distanceVar = disVar, ... bulkVar = 0, ... rateConstant = 0, ... otherVar = levVar, ... otherBulkVar = 0, ... otherRateConstant = 0)
>>> extVar = CellVariable(mesh = mesh, value = accVar.getInterfaceVar())
>>> from fipy.models.levelSet.advection.higherOrderAdvectionEquation \ ... import buildHigherOrderAdvectionEquation >>> advEq = buildHigherOrderAdvectionEquation(advectionCoeff = extVar)
>>> dt = 0.1
>>> for i in range(50): ... disVar.calcDistanceFunction() ... extVar.setValue(numerix.array(accVar.getInterfaceVar())) ... disVar.extendVariable(extVar) ... disVar.updateOld() ... advEq.solve(disVar, dt = dt) ... levEq.solve(levVar, dt = dt) ... accEq.solve(accVar, dt = dt)
>>> print (accVar >= -1e-10).all() True
Create a AdsorbingSurfactantEquation object.
Parameters : - surfactantVar: The SurfactantVariable to be solved for.
- distanceVar: The DistanceVariable that marks the interface.
- bulkVar: The value of the surfactantVar in the bulk.
- rateConstant: The adsorption rate of the surfactantVar.
- otherVar: Another SurfactantVariable with more surface affinity.
- otherBulkVar: The value of the otherVar in the bulk.
- otherRateConstant: The adsorption rate of the otherVar.
- consumptionCoeff: The rate that the surfactantVar is consumed during deposition.
- solve(var, boundaryConditions=(), solver=None, dt=1.0)¶
Builds and solves the AdsorbingSurfactantEquation‘s linear system once.
Parameters : - var: A SurfactantVariable to be solved for. Provides the initial condition, the old value and holds the solution on completion.
- solver: The iterative solver to be used to solve the linear system of equations.
- boundaryConditions: A tuple of boundaryConditions.
- dt: The time step size.
- sweep(var, solver=None, boundaryConditions=(), dt=1.0, underRelaxation=None, residualFn=None)¶
Builds and solves the AdsorbingSurfactantEquation‘s linear system once. This method also recalculates and returns the residual as well as applying under-relaxation.
Parameters : - var: The variable to be solved for. Provides the initial condition, the old value and holds the solution on completion.
- solver: The iterative solver to be used to solve the linear system of equations.
- boundaryConditions: A tuple of boundaryConditions.
- dt: The time step size.
- underRelaxation: Usually a value between 0 and 1 or None in the case of no under-relaxation
The convectionCoeff Module¶
The lines Module¶
The matplotlibSurfactantViewer Module¶
- class fipy.models.levelSet.surfactant.matplotlibSurfactantViewer.MatplotlibSurfactantViewer(distanceVar, surfactantVar=None, levelSetValue=0.0, title=None, smooth=0, zoomFactor=1.0, animate=False, limits={}, **kwlimits)¶
Bases: fipy.viewers.matplotlibViewer.matplotlibViewer._MatplotlibViewer
The MatplotlibSurfactantViewer creates a viewer with the Matplotlib python plotting package that displays a DistanceVariable.
Create a MatplotlibSurfactantViewer.
>>> from fipy import * >>> m = Grid2D(nx=100, ny=100) >>> x, y = m.getCellCenters() >>> v = CellVariable(mesh=m, value=x**2 + y**2 - 10**2) >>> s = CellVariable(mesh=m, value=sin(x / 10) * cos(y / 30)) >>> viewer = MatplotlibSurfactantViewer(distanceVar=v, surfactantVar=s) >>> for r in range(1,200): ... v.setValue(x**2 + y**2 - r**2) ... viewer.plot()
>>> from fipy import * >>> dx = 1. >>> dy = 1. >>> nx = 11 >>> ny = 11 >>> Lx = ny * dy >>> Ly = nx * dx >>> mesh = Grid2D(dx = dx, dy = dy, nx = nx, ny = ny) >>> # from fipy.models.levelSet.distanceFunction.distanceVariable import DistanceVariable >>> var = DistanceVariable(mesh = mesh, value = -1)
>>> x, y = mesh.getCellCenters()
>>> var.setValue(1, where=(x - Lx / 2.)**2 + (y - Ly / 2.)**2 < (Lx / 4.)**2) >>> var.calcDistanceFunction() >>> viewer = MatplotlibSurfactantViewer(var, smooth = 2) >>> viewer.plot() >>> viewer._promptForOpinion() >>> del viewer
>>> var = DistanceVariable(mesh = mesh, value = -1)
>>> var.setValue(1, where=(y > 2. * Ly / 3.) | ((x > Lx / 2.) & (y > Ly / 3.)) | ((y < Ly / 6.) & (x > Lx / 2))) >>> var.calcDistanceFunction() >>> viewer = MatplotlibSurfactantViewer(var) >>> viewer.plot() >>> viewer._promptForOpinion() >>> del viewer
>>> viewer = MatplotlibSurfactantViewer(var, smooth = 2) >>> viewer.plot() >>> viewer._promptForOpinion() >>> del viewer
Parameters : - distanceVar: a DistanceVariable object.
- levelSetValue: the value of the contour to be displayed
- title: displayed at the top of the Viewer window
- animate: whether to show only the initial condition and the
- limits: a dictionary with possible keys xmin, xmax, ymin, ymax, zmin, zmax, datamin, datamax. A 1D Viewer will only use xmin and xmax, a 2D viewer will also use ymin and ymax, and so on. All viewers will use datamin and datamax. Any limit set to a (default) value of None will autoscale. moving top boundary or to show all contours (Default)
The mayaviSurfactantViewer Module¶
- class fipy.models.levelSet.surfactant.mayaviSurfactantViewer.MayaviSurfactantViewer(distanceVar, surfactantVar=None, levelSetValue=0.0, title=None, smooth=0, zoomFactor=1.0, animate=False, limits={}, **kwlimits)¶
Bases: fipy.viewers.viewer._Viewer
The MayaviSurfactantViewer creates a viewer with the Mayavi python plotting package that displays a DistanceVariable.
Create a MayaviSurfactantViewer.
>>> from fipy import * >>> dx = 1. >>> dy = 1. >>> nx = 11 >>> ny = 11 >>> Lx = ny * dy >>> Ly = nx * dx >>> mesh = Grid2D(dx = dx, dy = dy, nx = nx, ny = ny) >>> # from fipy.models.levelSet.distanceFunction.distanceVariable import DistanceVariable >>> var = DistanceVariable(mesh = mesh, value = -1)
>>> x, y = mesh.getCellCenters()
>>> var.setValue(1, where=(x - Lx / 2.)**2 + (y - Ly / 2.)**2 < (Lx / 4.)**2) >>> var.calcDistanceFunction() >>> viewer = MayaviSurfactantViewer(var, smooth = 2) >>> viewer.plot() >>> viewer._promptForOpinion() >>> del viewer
>>> var = DistanceVariable(mesh = mesh, value = -1)
>>> var.setValue(1, where=(y > 2. * Ly / 3.) | ((x > Lx / 2.) & (y > Ly / 3.)) | ((y < Ly / 6.) & (x > Lx / 2))) >>> var.calcDistanceFunction() >>> viewer = MayaviSurfactantViewer(var) >>> viewer.plot() >>> viewer._promptForOpinion() >>> del viewer
>>> viewer = MayaviSurfactantViewer(var, smooth = 2) >>> viewer.plot() >>> viewer._promptForOpinion() >>> del viewer
Parameters : - distanceVar: a DistanceVariable object.
- levelSetValue: the value of the contour to be displayed
- title: displayed at the top of the Viewer window
- animate: whether to show only the initial condition and the
- limits: a dictionary with possible keys xmin, xmax, ymin, ymax, zmin, zmax, datamin, datamax. A 1D Viewer will only use xmin and xmax, a 2D viewer will also use ymin and ymax, and so on. All viewers will use datamin and datamax. Any limit set to a (default) value of None will autoscale. moving top boundary or to show all contours (Default)
- plot(filename=None)¶
The surfactantBulkDiffusionEquation Module¶
- fipy.models.levelSet.surfactant.surfactantBulkDiffusionEquation.buildSurfactantBulkDiffusionEquation(bulkVar=None, distanceVar=None, surfactantVar=None, otherSurfactantVar=None, diffusionCoeff=None, transientCoeff=1.0, rateConstant=None)¶
The buildSurfactantBulkDiffusionEquation function returns a bulk diffusion of a species with a source term for the jump from the bulk to an interface. The governing equation is given by,
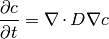
where,
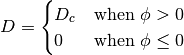
The jump condition at the interface is defined by Langmuir adsorption. Langmuir adsorption essentially states that the ability for a species to jump from an electrolyte to an interface is proportional to the concentration in the electrolyte, available site density and a jump coefficient. The boundary condition at the interface is given by
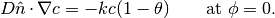
Parameters : - bulkVar: The bulk surfactant concentration variable.
- distanceVar: A DistanceVariable object
- surfactantVar: A SurfactantVariable object
- otherSurfactantVar: Any other surfactants that may remove this one.
- diffusionCoeff: A float or a FaceVariable.
- transientCoeff: In general 1 is used.
- rateConstant: The adsorption coefficient.
The surfactantEquation Module¶
- class fipy.models.levelSet.surfactant.surfactantEquation.SurfactantEquation(distanceVar=None)¶
A SurfactantEquation aims to evolve a surfactant on an interface defined by the zero level set of the distanceVar. The method should completely conserve the total coverage of surfactant. The surfactant is only in the cells immediately in front of the advancing interface. The method only works for a positive velocity as it stands.
Creates a SurfactantEquation object.
Parameters : - distanceVar: The DistanceVariable that marks the interface.
- solve(var, boundaryConditions=(), solver=None, dt=1.0)¶
Builds and solves the SurfactantEquation‘s linear system once.
Parameters : - var: A SurfactantVariable to be solved for. Provides the initial condition, the old value and holds the solution on completion.
- solver: The iterative solver to be used to solve the linear system of equations.
- boundaryConditions: A tuple of boundaryConditions.
- dt: The time step size.
- sweep(var, solver=None, boundaryConditions=(), dt=1.0, underRelaxation=None, residualFn=None)¶
Builds and solves the Term‘s linear system once. This method also recalculates and returns the residual as well as applying under-relaxation.
Parameters : - var: The variable to be solved for. Provides the initial condition, the old value and holds the solution on completion.
- solver: The iterative solver to be used to solve the linear system of equations.
- boundaryConditions: A tuple of boundaryConditions.
- dt: The time step size.
- underRelaxation: Usually a value between 0 and 1 or None in the case of no under-relaxation
The surfactantVariable Module¶
- class fipy.models.levelSet.surfactant.surfactantVariable.SurfactantVariable(value=0.0, distanceVar=None, name='surfactant variable', hasOld=False)¶
Bases: fipy.variables.cellVariable.CellVariable
The SurfactantVariable maintains a conserved volumetric concentration on cells adjacent to, but in front of, the interface. The value argument corresponds to the initial concentration of surfactant on the interface (moles divided by area). The value held by the SurfactantVariable is actually a volume density (moles divided by volume).
A simple 1D test:
>>> from fipy.meshes.grid1D import Grid1D >>> mesh = Grid1D(dx = 1., nx = 4) >>> from fipy.models.levelSet.distanceFunction.distanceVariable \ ... import DistanceVariable >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (-1.5, -0.5, 0.5, 941.5)) >>> surfactantVariable = SurfactantVariable(value = 1, ... distanceVar = distanceVariable) >>> print numerix.allclose(surfactantVariable, (0, 0., 1., 0)) 1
A 2D test case:
>>> from fipy.meshes.grid2D import Grid2D >>> mesh = Grid2D(dx = 1., dy = 1., nx = 3, ny = 3) >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (1.5, 0.5, 1.5, ... 0.5,-0.5, 0.5, ... 1.5, 0.5, 1.5)) >>> surfactantVariable = SurfactantVariable(value = 1, ... distanceVar = distanceVariable) >>> print numerix.allclose(surfactantVariable, (0, 1, 0, 1, 0, 1, 0, 1, 0)) 1
Another 2D test case:
>>> mesh = Grid2D(dx = .5, dy = .5, nx = 2, ny = 2) >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (-0.5, 0.5, 0.5, 1.5)) >>> surfactantVariable = SurfactantVariable(value = 1, ... distanceVar = distanceVariable) >>> print numerix.allclose(surfactantVariable, ... (0, numerix.sqrt(2), numerix.sqrt(2), 0)) 1
Parameters : - value: The initial value.
- distanceVar: A DistanceVariable object.
- name: The name of the variable.
- copy()¶
- getInterfaceVar()¶
Returns the SurfactantVariable rendered as an _InterfaceSurfactantVariable which evaluates the surfactant concentration as an area concentration the interface rather than a volumetric concentration.

 FiPy
FiPy