fipy.variables package¶
Submodules¶
fipy.variables.addOverFacesVariable module¶
fipy.variables.arithmeticCellToFaceVariable module¶
fipy.variables.betaNoiseVariable module¶
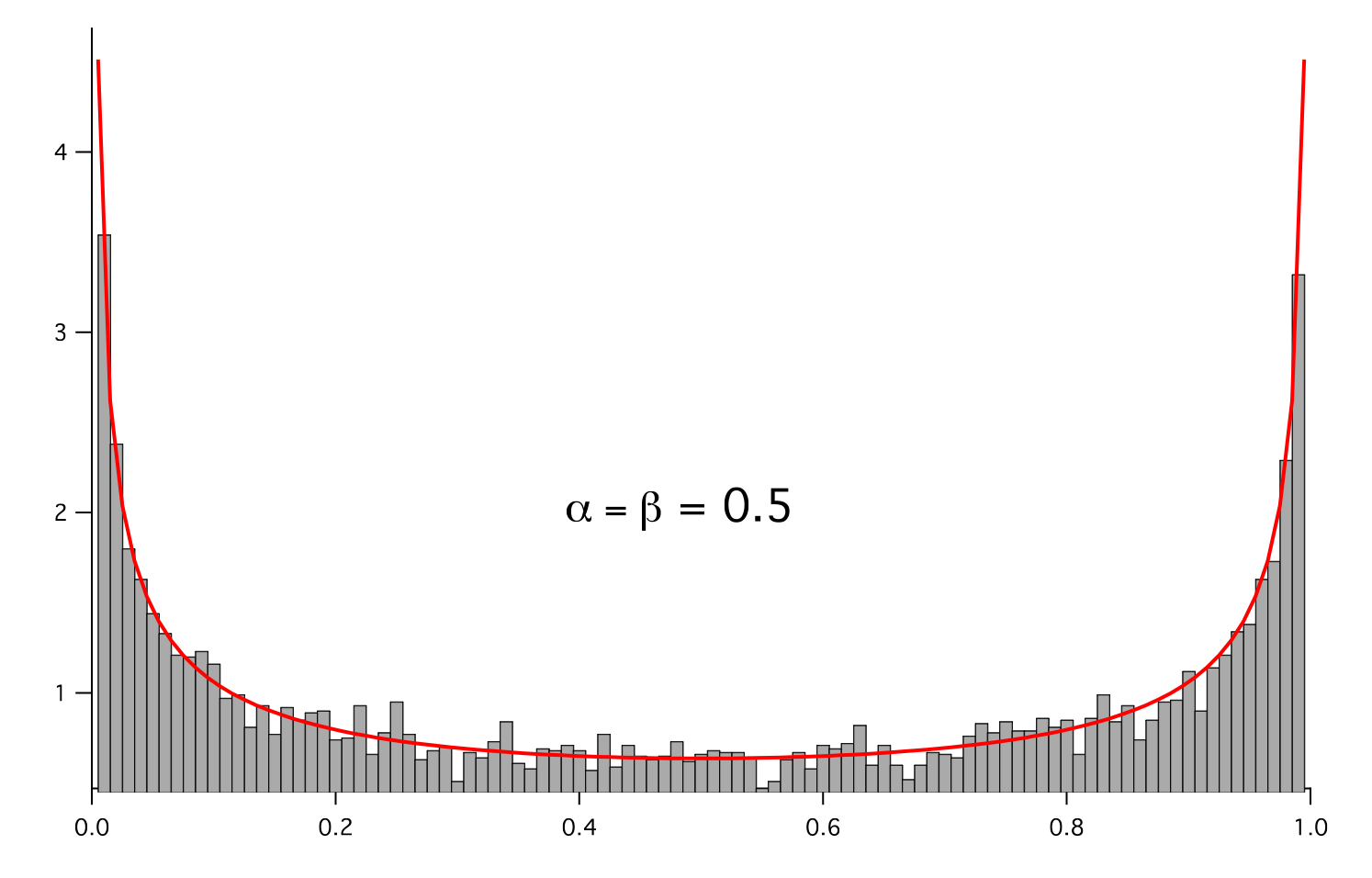
- class fipy.variables.betaNoiseVariable.BetaNoiseVariable(*args, **kwds)¶
Bases:
NoiseVariableRepresents a beta distribution of random numbers with the probability distribution
with a shape parameter
, a rate parameter
, and
.
Seed the random module for the sake of deterministic test results.
>>> from fipy import numerix >>> numerix.random.seed(1)
We generate noise on a uniform Cartesian mesh
>>> from fipy.variables.variable import Variable >>> alpha = Variable() >>> beta = Variable() >>> from fipy.meshes import Grid2D >>> noise = BetaNoiseVariable(mesh = Grid2D(nx = 100, ny = 100), alpha = alpha, beta = beta)
We histogram the root-volume-weighted noise distribution
>>> from fipy.variables.histogramVariable import HistogramVariable >>> histogram = HistogramVariable(distribution = noise, dx = 0.01, nx = 100)
and compare to a Gaussian distribution
>>> from fipy.variables.cellVariable import CellVariable >>> betadist = CellVariable(mesh = histogram.mesh) >>> x = CellVariable(mesh=histogram.mesh, value=histogram.mesh.cellCenters[0]) >>> from scipy.special import gamma as Gamma >>> betadist = ((Gamma(alpha + beta) / (Gamma(alpha) * Gamma(beta))) ... * x**(alpha - 1) * (1 - x)**(beta - 1))
>>> if __name__ == '__main__': ... from fipy import Viewer ... viewer = Viewer(vars=noise, datamin=0, datamax=1) ... histoplot = Viewer(vars=(histogram, betadist), ... datamin=0, datamax=1.5)
>>> from fipy.tools.numerix import arange
>>> for a in arange(0.5, 5, 0.5): ... alpha.value = a ... for b in arange(0.5, 5, 0.5): ... beta.value = b ... if __name__ == '__main__': ... import sys ... print("alpha: %g, beta: %g" % (alpha, beta), file=sys.stderr) ... viewer.plot() ... histoplot.plot()
>>> print(abs(noise.faceGrad.divergence.cellVolumeAverage) < 5e-15) 1

- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Copy the value of the NoiseVariable to a static CellVariable.
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- parallelRandom()¶
- put(indices, value)¶
- random()¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- scramble()¶
Generate a new random distribution.
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.binaryOperatorVariable module¶
fipy.variables.cellToFaceVariable module¶
fipy.variables.cellVariable module¶
- class fipy.variables.cellVariable.CellVariable(*args, **kwds)¶
Bases:
_MeshVariableRepresents the field of values of a variable on a Mesh.
A CellVariable can be
pickledto persistent storage (disk) for later use:>>> from fipy.meshes import Grid2D >>> mesh = Grid2D(dx = 1., dy = 1., nx = 10, ny = 10)
>>> var = CellVariable(mesh = mesh, value = 1., hasOld = 1, name = 'test') >>> x, y = mesh.cellCenters >>> var.value = (x * y)
>>> from fipy.tools import dump >>> (f, filename) = dump.write(var, extension = '.gz') >>> unPickledVar = dump.read(filename, f)
>>> print(var.allclose(unPickledVar, atol = 1e-10, rtol = 1e-10)) 1
- Parameters:
mesh (Mesh) – the mesh that defines the geometry of this Variable
name (str) – the user-readable name of the Variable
value (float or array_like) – the initial value
rank (int) – the rank (number of dimensions) of each element of this Variable. Default: 0
elementshape (
tupleofint) – the shape of each element of this variable Default: rank * (mesh.dim,)unit (str or PhysicalUnit) – The physical units of the variable
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.constant module¶
fipy.variables.constraintMask module¶
fipy.variables.coupledCellVariable module¶
fipy.variables.distanceVariable module¶
- class fipy.variables.distanceVariable.DistanceVariable(*args, **kwds)¶
Bases:
CellVariableA DistanceVariable object calculates
so it satisfies,
using the fast marching method with an initial condition defined by the zero level set. The solution can either be first or second order.
Here we will define a few test cases. Firstly a 1D test case
>>> from fipy.meshes import Grid1D >>> from fipy.tools import serialComm >>> mesh = Grid1D(dx = .5, nx = 8, communicator=serialComm) >>> from .distanceVariable import DistanceVariable >>> var = DistanceVariable(mesh = mesh, value = (-1., -1., -1., -1., 1., 1., 1., 1.)) >>> var.calcDistanceFunction() >>> answer = (-1.75, -1.25, -.75, -0.25, 0.25, 0.75, 1.25, 1.75) >>> print(var.allclose(answer)) 1
A 1D test case with very small dimensions.
>>> dx = 1e-10 >>> mesh = Grid1D(dx = dx, nx = 8, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., -1., -1., -1., 1., 1., 1., 1.)) >>> var.calcDistanceFunction() >>> answer = numerix.arange(8) * dx - 3.5 * dx >>> print(var.allclose(answer)) 1
A 2D test case to test _calcTrialValue for a pathological case.
>>> dx = 1. >>> dy = 2. >>> from fipy.meshes import Grid2D >>> mesh = Grid2D(dx = dx, dy = dy, nx = 2, ny = 3, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., 1., 1., 1., -1., 1.))
>>> var.calcDistanceFunction() >>> vbl = -dx * dy / numerix.sqrt(dx**2 + dy**2) / 2. >>> vbr = dx / 2 >>> vml = dy / 2. >>> crossProd = dx * dy >>> dsq = dx**2 + dy**2 >>> top = vbr * dx**2 + vml * dy**2 >>> sqrt = crossProd**2 *(dsq - (vbr - vml)**2) >>> sqrt = numerix.sqrt(max(sqrt, 0)) >>> vmr = (top + sqrt) / dsq >>> answer = (vbl, vbr, vml, vmr, vbl, vbr) >>> print(var.allclose(answer)) 1
The extendVariable method solves the following equation for a given extensionVariable.
using the fast marching method with an initial condition defined at the zero level set.
>>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(dx = 1., dy = 1., nx = 2, ny = 2, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., 1., 1., 1.)) >>> var.calcDistanceFunction() >>> extensionVar = CellVariable(mesh = mesh, value = (-1, .5, 2, -1)) >>> tmp = 1 / numerix.sqrt(2) >>> print(var.allclose((-tmp / 2, 0.5, 0.5, 0.5 + tmp))) 1 >>> var.extendVariable(extensionVar, order=1) >>> print(extensionVar.allclose((1.25, .5, 2, 1.25))) 1 >>> mesh = Grid2D(dx = 1., dy = 1., nx = 3, ny = 3, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., 1., 1., ... 1., 1., 1., ... 1., 1., 1.)) >>> var.calcDistanceFunction(order=1) >>> extensionVar = CellVariable(mesh = mesh, value = (-1., .5, -1., ... 2., -1., -1., ... -1., -1., -1.))
>>> v1 = 0.5 + tmp >>> v2 = 1.5 >>> tmp1 = (v1 + v2) / 2 + numerix.sqrt(2. - (v1 - v2)**2) / 2 >>> tmp2 = tmp1 + 1 / numerix.sqrt(2) >>> print(var.allclose((-tmp / 2, 0.5, 1.5, 0.5, 0.5 + tmp, ... tmp1, 1.5, tmp1, tmp2))) 1 >>> answer = (1.25, .5, .5, 2, 1.25, 0.9544, 2, 1.5456, 1.25) >>> var.extendVariable(extensionVar, order=1) >>> print(extensionVar.allclose(answer, rtol = 1e-4)) 1
Test case for a bug that occurs when initializing the distance variable at the interface. Currently it is assumed that adjacent cells that are opposite sign neighbors have perpendicular normal vectors. In fact the two closest cells could have opposite normals.
>>> mesh = Grid1D(dx = 1., nx = 3, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., 1., -1.)) >>> var.calcDistanceFunction() >>> print(var.allclose((-0.5, 0.5, -0.5))) 1
Testing second order. This example failed with Scikit-fmm.
>>> mesh = Grid2D(dx = 1., dy = 1., nx = 4, ny = 4, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., -1., 1., 1., ... -1., -1., 1., 1., ... 1., 1., 1., 1., ... 1, 1, 1, 1)) >>> var.calcDistanceFunction(order=2) >>> answer = [-1.30473785, -0.5, 0.5, 1.49923009, ... -0.5, -0.35355339, 0.5, 1.45118446, ... 0.5, 0.5, 0.97140452, 1.76215286, ... 1.49923009, 1.45118446, 1.76215286, 2.33721352] >>> print(numerix.allclose(var, answer, rtol=1e-9)) True
** A test for a bug in both LSMLIB and Scikit-fmm **
The following test gives different result depending on whether LSMLIB or Scikit-fmm is used. There is a deeper problem that is related to this issue. When a value becomes “known” after previously being a “trial” value it updates its neighbors’ values. In a second order scheme the neighbors one step away also need to be updated (if the in between cell is “known” and the far cell is a “trial” cell), but are not in either package. By luck (due to trial values having the same value), the values calculated in Scikit-fmm for the following example are correct although an example that didn’t work for Scikit-fmm could also be constructed.
>>> mesh = Grid2D(dx = 1., dy = 1., nx = 4, ny = 4, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., -1., -1., -1., ... 1., 1., -1., -1., ... 1., 1., -1., -1., ... 1., 1., -1., -1.)) >>> var.calcDistanceFunction(order=2) >>> var.calcDistanceFunction(order=2) >>> answer = [-0.5, -0.58578644, -1.08578644, -1.85136395, ... 0.5, 0.29289322, -0.58578644, -1.54389939, ... 1.30473785, 0.5, -0.5, -1.5, ... 1.49547948, 0.5, -0.5, -1.5]
The 3rd and 7th element are different for LSMLIB. This is because the 15th element is not “known” when the “trial” value for the 7th element is calculated. Scikit-fmm calculates the values in a slightly different order so gets a seemingly better answer, but this is just chance.
>>> print(numerix.allclose(var, answer, rtol=1e-9)) True
Creates a distanceVariable object.
- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- calcDistanceFunction(order=2)¶
Calculates the distanceVariable as a distance function.
- Parameters:
order ({1, 2}) – The order of accuracy for the distance function calculation
- property cellInterfaceAreas¶
Returns the length of the interface that crosses the cell
A simple 1D test:
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(dx = 1., nx = 4) >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (-1.5, -0.5, 0.5, 1.5)) >>> answer = CellVariable(mesh=mesh, value=(0, 0., 1., 0)) >>> print(numerix.allclose(distanceVariable.cellInterfaceAreas, ... answer)) True
A 2D test case:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(dx = 1., dy = 1., nx = 3, ny = 3) >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (1.5, 0.5, 1.5, ... 0.5, -0.5, 0.5, ... 1.5, 0.5, 1.5)) >>> answer = CellVariable(mesh=mesh, ... value=(0, 1, 0, 1, 0, 1, 0, 1, 0)) >>> print(numerix.allclose(distanceVariable.cellInterfaceAreas, answer)) True
Another 2D test case:
>>> mesh = Grid2D(dx = .5, dy = .5, nx = 2, ny = 2) >>> from fipy.variables.cellVariable import CellVariable >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (-0.5, 0.5, 0.5, 1.5)) >>> answer = CellVariable(mesh=mesh, ... value=(0, numerix.sqrt(2) / 4, numerix.sqrt(2) / 4, 0)) >>> print(numerix.allclose(distanceVariable.cellInterfaceAreas, ... answer)) True
Test to check that the circumference of a circle is, in fact,
.
>>> mesh = Grid2D(dx = 0.05, dy = 0.05, nx = 20, ny = 20) >>> r = 0.25 >>> x, y = mesh.cellCenters >>> rad = numerix.sqrt((x - .5)**2 + (y - .5)**2) - r >>> distanceVariable = DistanceVariable(mesh = mesh, value = rad) >>> print(numerix.allclose(distanceVariable.cellInterfaceAreas.sum(), 1.57984690073)) 1
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- extendVariable(extensionVariable, order=2)¶
Calculates the extension of extensionVariable from the zero level set.
- Parameters:
extensionVariable (CellVariable) – The variable to extend from the zero level set.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getLSMshape()¶
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.exponentialNoiseVariable module¶
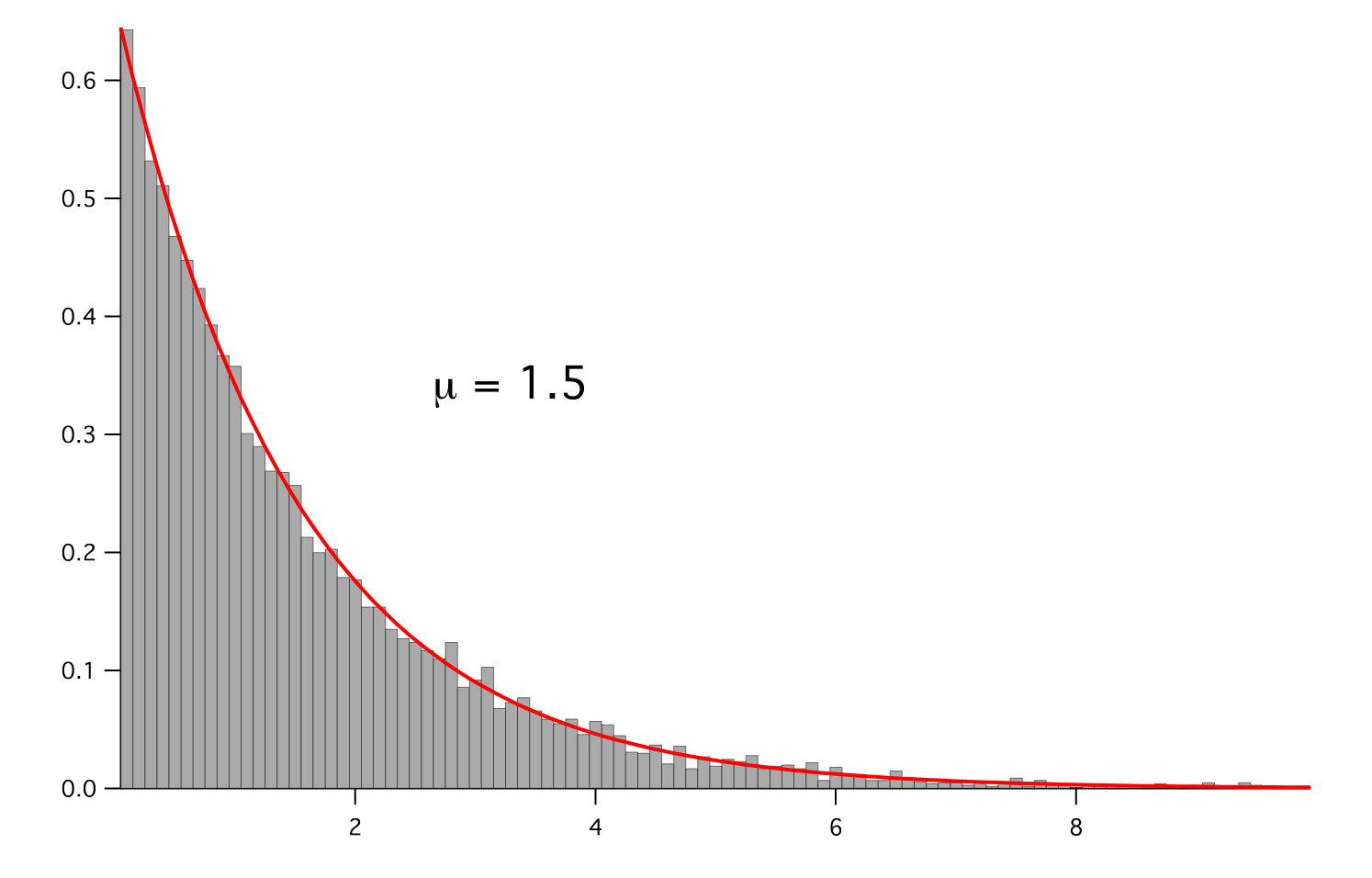
- class fipy.variables.exponentialNoiseVariable.ExponentialNoiseVariable(*args, **kwds)¶
Bases:
NoiseVariableRepresents an exponential distribution of random numbers with the probability distribution
with a mean parameter
.
Seed the random module for the sake of deterministic test results.
>>> from fipy import numerix >>> numerix.random.seed(1)
We generate noise on a uniform Cartesian mesh
>>> from fipy.variables.variable import Variable >>> mean = Variable() >>> from fipy.meshes import Grid2D >>> noise = ExponentialNoiseVariable(mesh = Grid2D(nx = 100, ny = 100), mean = mean)
We histogram the root-volume-weighted noise distribution
>>> from fipy.variables.histogramVariable import HistogramVariable >>> histogram = HistogramVariable(distribution = noise, dx = 0.1, nx = 100)
and compare to a Gaussian distribution
>>> from fipy.variables.cellVariable import CellVariable >>> expdist = CellVariable(mesh = histogram.mesh) >>> x = histogram.mesh.cellCenters[0]
>>> if __name__ == '__main__': ... from fipy import Viewer ... viewer = Viewer(vars=noise, datamin=0, datamax=5) ... histoplot = Viewer(vars=(histogram, expdist), ... datamin=0, datamax=1.5)
>>> from fipy.tools.numerix import arange, exp
>>> for mu in arange(0.5, 3, 0.5): ... mean.value = (mu) ... expdist.value = ((1/mean)*exp(-x/mean)) ... if __name__ == '__main__': ... import sys ... print("mean: %g" % mean, file=sys.stderr) ... viewer.plot() ... histoplot.plot()
>>> print(abs(noise.faceGrad.divergence.cellVolumeAverage) < 5e-15) 1
- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Copy the value of the NoiseVariable to a static CellVariable.
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- parallelRandom()¶
- put(indices, value)¶
- random()¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- scramble()¶
Generate a new random distribution.
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.faceGradContributionsVariable module¶
fipy.variables.faceGradVariable module¶
fipy.variables.faceVariable module¶
- class fipy.variables.faceVariable.FaceVariable(*args, **kwds)¶
Bases:
_MeshVariable- Parameters:
mesh (Mesh) – the mesh that defines the geometry of this Variable
name (str) – the user-readable name of the Variable
value (float or array_like) – the initial value
rank (int) – the rank (number of dimensions) of each element of this Variable. Default: 0
elementshape (
tupleofint) – the shape of each element of this variable Default: rank * (mesh.dim,)unit (str or PhysicalUnit) – The physical units of the variable
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__()¶
“Evaluate” the Variable and return its value
>>> a = Variable(value=3) >>> print(a()) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b() 7
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe _MeshVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new Variable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- cacheMe(recursive=False)¶
- constrain(value, where=None)¶
Constrain the Variable to have a value at an index or mask location specified by where.
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> print(v) [2 1 2 3] >>> v[:] = 10 >>> print(v) [ 2 10 10 10] >>> v.constrain(5, numerix.array((False, False, True, False))) >>> print(v) [ 2 10 5 10] >>> v[:] = 6 >>> print(v) [2 6 5 6] >>> v.constrain(8) >>> print(v) [8 8 8 8] >>> v[:] = 10 >>> print(v) [8 8 8 8] >>> del v.constraints[2] >>> print(v) [ 2 10 5 10]
>>> from fipy.variables.cellVariable import CellVariable >>> from fipy.meshes import Grid2D >>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v = CellVariable(mesh=m, rank=1, value=(x, y)) >>> v.constrain(((0.,), (-1.,)), where=m.facesLeft) >>> print(v.faceValue) [[ 0.5 1.5 0.5 1.5 0.5 1.5 0. 1. 1.5 0. 1. 1.5] [ 0.5 0.5 1. 1. 1.5 1.5 -1. 0.5 0.5 -1. 1.5 1.5]]
- Parameters:
value (float or array_like) – The value of the constraint
where (array_like of
bool) – The constraint mask or index specifying the location of the constraint
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- property divergence¶
the divergence of self,
,
- Returns:
divergence – one rank lower than self
- Return type:
Examples
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=3, ny=2) >>> from builtins import range >>> var = CellVariable(mesh=mesh, value=list(range(3*2))) >>> print(var.faceGrad.divergence) [ 4. 3. 2. -2. -3. -4.]
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property name¶
- property numericValue¶
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> v[:] = 10 >>> from fipy.boundaryConditions.constraint import Constraint >>> c1 = Constraint(5, numerix.array((False, False, True, False))) >>> v.constrain(c1) >>> v[:] = 6 >>> v.constrain(8) >>> v[:] = 10 >>> del v.constraints[2] >>> v.release(constraint=c1) >>> print(v) [ 2 10 10 10]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.gammaNoiseVariable module¶
- class fipy.variables.gammaNoiseVariable.GammaNoiseVariable(*args, **kwds)¶
Bases:
NoiseVariableRepresents a gamma distribution of random numbers with the probability distribution
with a shape parameter
, a rate parameter
, and
.
Seed the random module for the sake of deterministic test results.
>>> from fipy import numerix >>> numerix.random.seed(1)
We generate noise on a uniform Cartesian mesh
>>> from fipy.variables.variable import Variable >>> alpha = Variable() >>> beta = Variable() >>> from fipy.meshes import Grid2D >>> noise = GammaNoiseVariable(mesh = Grid2D(nx = 100, ny = 100), shape = alpha, rate = beta)
We histogram the root-volume-weighted noise distribution
>>> from fipy.variables.histogramVariable import HistogramVariable >>> histogram = HistogramVariable(distribution = noise, dx = 0.1, nx = 300)
and compare to a Gaussian distribution
>>> from fipy.variables.cellVariable import CellVariable >>> x = CellVariable(mesh=histogram.mesh, value=histogram.mesh.cellCenters[0]) >>> from scipy.special import gamma as Gamma >>> from fipy.tools.numerix import exp >>> gammadist = (x**(alpha - 1) * (beta**alpha * exp(-beta * x)) / Gamma(alpha))
>>> if __name__ == '__main__': ... from fipy import Viewer ... viewer = Viewer(vars=noise, datamin=0, datamax=30) ... histoplot = Viewer(vars=(histogram, gammadist), ... datamin=0, datamax=1)
>>> from fipy.tools.numerix import arange
>>> for shape in arange(1, 8, 1): ... alpha.value = shape ... for rate in arange(0.5, 2.5, 0.5): ... beta.value = rate ... if __name__ == '__main__': ... import sys ... print("alpha: %g, beta: %g" % (alpha, beta), file=sys.stderr) ... viewer.plot() ... histoplot.plot()
>>> print(abs(noise.faceGrad.divergence.cellVolumeAverage) < 5e-15) 1

- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Copy the value of the NoiseVariable to a static CellVariable.
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- parallelRandom()¶
- put(indices, value)¶
- random()¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- scramble()¶
Generate a new random distribution.
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.gaussCellGradVariable module¶
fipy.variables.gaussianNoiseVariable module¶
- class fipy.variables.gaussianNoiseVariable.GaussianNoiseVariable(*args, **kwds)¶
Bases:
NoiseVariableRepresents a normal (Gaussian) distribution of random numbers with mean
and variance
, which has the probability distribution
For example, the variance of thermal noise that is uncorrelated in space and time is often expressed as
which can be obtained with:
sigmaSqrd = Mobility * kBoltzmann * Temperature / (mesh.cellVolumes * timeStep) GaussianNoiseVariable(mesh = mesh, variance = sigmaSqrd)
Note
If the time step will change as the simulation progresses, either through use of an adaptive iterator or by making manual changes at different stages, remember to declare timeStep as a Variable and to change its value with its setValue() method.
>>> import sys >>> from fipy.tools.numerix import *
>>> mean = 0. >>> variance = 4.
Seed the random module for the sake of deterministic test results.
>>> from fipy import numerix >>> numerix.random.seed(3)
We generate noise on a non-uniform Cartesian mesh with cell dimensions of
and
.
>>> from fipy.meshes import Grid2D >>> mesh = Grid2D(dx = arange(0.1, 5., 0.1)**2, dy = arange(0.1, 3., 0.1)**3) >>> from fipy.variables.cellVariable import CellVariable >>> volumes = CellVariable(mesh=mesh, value=mesh.cellVolumes) >>> noise = GaussianNoiseVariable(mesh = mesh, mean = mean, ... variance = variance / volumes)
We histogram the root-volume-weighted noise distribution
>>> from fipy.variables.histogramVariable import HistogramVariable >>> histogram = HistogramVariable(distribution = noise * sqrt(volumes), ... dx = 0.1, nx = 600, offset = -30)
and compare to a Gaussian distribution
>>> gauss = CellVariable(mesh = histogram.mesh) >>> x = histogram.mesh.cellCenters[0] >>> gauss.value = ((1/(sqrt(variance * 2 * pi))) * exp(-(x - mean)**2 / (2 * variance)))
>>> if __name__ == '__main__': ... from fipy.viewers import Viewer ... viewer = Viewer(vars=noise, ... datamin=-5, datamax=5) ... histoplot = Viewer(vars=(histogram, gauss))
>>> from builtins import range >>> for i in range(10): ... noise.scramble() ... if __name__ == '__main__': ... viewer.plot() ... histoplot.plot()
>>> print(abs(noise.faceGrad.divergence.cellVolumeAverage) < 5e-15) 1
Note that the noise exhibits larger amplitude in the small cells than in the large ones
but that the root-volume-weighted histogram is Gaussian.
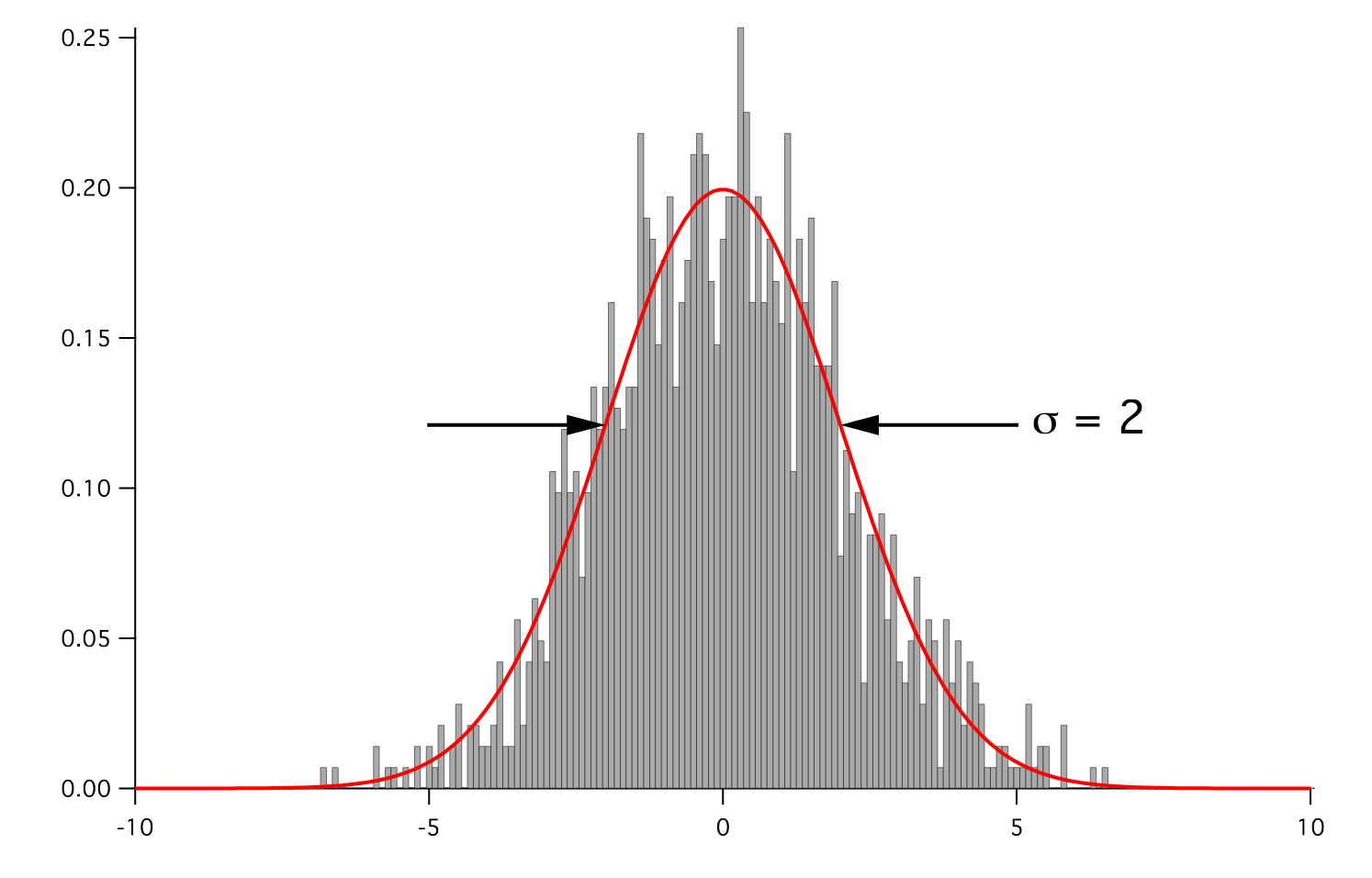
- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Copy the value of the NoiseVariable to a static CellVariable.
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- parallelRandom()¶
- put(indices, value)¶
- random()¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- scramble()¶
Generate a new random distribution.
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.harmonicCellToFaceVariable module¶
fipy.variables.histogramVariable module¶
- class fipy.variables.histogramVariable.HistogramVariable(*args, **kwds)¶
Bases:
CellVariableProduces a histogram of the values of the supplied distribution.
- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.interfaceAreaVariable module¶
fipy.variables.interfaceFlagVariable module¶
fipy.variables.leastSquaresCellGradVariable module¶
fipy.variables.levelSetDiffusionVariable module¶
fipy.variables.meshVariable module¶
fipy.variables.minmodCellToFaceVariable module¶
fipy.variables.modCellGradVariable module¶
fipy.variables.modCellToFaceVariable module¶
fipy.variables.modFaceGradVariable module¶
fipy.variables.modPhysicalField module¶
fipy.variables.modularVariable module¶
- class fipy.variables.modularVariable.ModularVariable(*args, **kwds)¶
Bases:
CellVariableThe ModularVariable defines a variable that exists on the circle between
and
The following examples show how ModularVariable works. When subtracting the answer wraps back around the circle.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.tools import numerix >>> pi = numerix.pi >>> v1 = ModularVariable(mesh = mesh, value = (2*pi/3, -2*pi/3)) >>> v2 = ModularVariable(mesh = mesh, value = -2*pi/3) >>> print(numerix.allclose(v2 - v1, (2*pi/3, 0))) 1
Obtaining the arithmetic face value.
>>> print(numerix.allclose(v1.arithmeticFaceValue, (2*pi/3, pi, -2*pi/3))) 1
Obtaining the gradient.
>>> print(numerix.allclose(v1.grad, ((pi/3, pi/3),))) 1
Obtaining the gradient at the faces.
>>> print(numerix.allclose(v1.faceGrad, ((0, 2*pi/3, 0),))) 1
Obtaining the gradient at the faces but without modular arithmetic.
>>> print(numerix.allclose(v1.faceGradNoMod, ((0, -4*pi/3, 0),))) 1
- Parameters:
mesh (Mesh) – the mesh that defines the geometry of this Variable
name (str) – the user-readable name of the Variable
value (float or array_like) – the initial value
rank (int) – the rank (number of dimensions) of each element of this Variable. Default: 0
elementshape (
tupleofint) – the shape of each element of this variable Default: rank * (mesh.dim,)unit (str or PhysicalUnit) – The physical units of the variable
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
Adjusted for a ModularVariable
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable (second-order gradient). Adjusted for a ModularVariable
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceGradNoMod¶
Return
as a rank-1 FaceVariable (second-order gradient). Not adjusted for a ModularVariable
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient). Adjusted for a ModularVariable
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values. Test case due to bug.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 1) >>> var = ModularVariable(mesh=mesh, value=1., hasOld=1) >>> var.updateOld() >>> var[:] = 2 >>> answer = CellVariable(mesh=mesh, value=1.) >>> print(var.old.allclose(answer)) True
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.noiseVariable module¶
- class fipy.variables.noiseVariable.NoiseVariable(*args, **kwds)¶
Bases:
CellVariableAttention
This class is abstract. Always create one of its subclasses.
A generic base class for sources of noise distributed over the cells of a mesh.
In the event that the noise should be conserved, use:
<Specific>NoiseVariable(...).faceGrad.divergence
The seed() and get_seed() functions of the fipy.tools.numerix.random module can be set and query the random number generated used by all NoiseVariable objects.
- Parameters:
mesh (Mesh) – the mesh that defines the geometry of this Variable
name (str) – the user-readable name of the Variable
value (float or array_like) – the initial value
rank (int) – the rank (number of dimensions) of each element of this Variable. Default: 0
elementshape (
tupleofint) – the shape of each element of this variable Default: rank * (mesh.dim,)unit (str or PhysicalUnit) – The physical units of the variable
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Copy the value of the NoiseVariable to a static CellVariable.
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- parallelRandom()¶
- put(indices, value)¶
- random()¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- scramble()¶
Generate a new random distribution.
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.operatorVariable module¶
fipy.variables.scharfetterGummelFaceVariable module¶
- class fipy.variables.scharfetterGummelFaceVariable.ScharfetterGummelFaceVariable(*args, **kwds)¶
Bases:
_CellToFaceVariable- Parameters:
mesh (Mesh) – the mesh that defines the geometry of this Variable
name (str) – the user-readable name of the Variable
value (float or array_like) – the initial value
rank (int) – the rank (number of dimensions) of each element of this Variable. Default: 0
elementshape (
tupleofint) – the shape of each element of this variable Default: rank * (mesh.dim,)unit (str or PhysicalUnit) – The physical units of the variable
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__()¶
“Evaluate” the Variable and return its value
>>> a = Variable(value=3) >>> print(a()) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b() 7
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe _MeshVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new Variable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- cacheMe(recursive=False)¶
- constrain(value, where=None)¶
Constrain the Variable to have a value at an index or mask location specified by where.
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> print(v) [2 1 2 3] >>> v[:] = 10 >>> print(v) [ 2 10 10 10] >>> v.constrain(5, numerix.array((False, False, True, False))) >>> print(v) [ 2 10 5 10] >>> v[:] = 6 >>> print(v) [2 6 5 6] >>> v.constrain(8) >>> print(v) [8 8 8 8] >>> v[:] = 10 >>> print(v) [8 8 8 8] >>> del v.constraints[2] >>> print(v) [ 2 10 5 10]
>>> from fipy.variables.cellVariable import CellVariable >>> from fipy.meshes import Grid2D >>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v = CellVariable(mesh=m, rank=1, value=(x, y)) >>> v.constrain(((0.,), (-1.,)), where=m.facesLeft) >>> print(v.faceValue) [[ 0.5 1.5 0.5 1.5 0.5 1.5 0. 1. 1.5 0. 1. 1.5] [ 0.5 0.5 1. 1. 1.5 1.5 -1. 0.5 0.5 -1. 1.5 1.5]]
- Parameters:
value (float or array_like) – The value of the constraint
where (array_like of
bool) – The constraint mask or index specifying the location of the constraint
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- property divergence¶
the divergence of self,
,
- Returns:
divergence – one rank lower than self
- Return type:
Examples
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=3, ny=2) >>> from builtins import range >>> var = CellVariable(mesh=mesh, value=list(range(3*2))) >>> print(var.faceGrad.divergence) [ 4. 3. 2. -2. -3. -4.]
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property name¶
- property numericValue¶
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c0 = Constraint(0., where=m.facesLeft) >>> v.constrain(c0) >>> c1 = Constraint(3., where=m.facesRight) >>> v.faceValue.constrain(c1) >>> print(v.faceValue) [ 0. 1. 2. 3.] >>> v.faceValue.release(constraint=c0) >>> print(v.faceValue) [ 0.5 1. 2. 3. ] >>> v.faceValue.release(constraint=c1) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.surfactantConvectionVariable module¶
- class fipy.variables.surfactantConvectionVariable.SurfactantConvectionVariable(*args, **kwds)¶
Bases:
FaceVariableConvection coefficient for the ConservativeSurfactantEquation. The coefficient only has a value for a negative distanceVar.
Simple one dimensional test:
>>> from fipy.variables.cellVariable import CellVariable >>> from fipy.meshes import Grid2D >>> mesh = Grid2D(nx = 3, ny = 1, dx = 1., dy = 1.) >>> from fipy.variables.distanceVariable import DistanceVariable >>> distanceVar = DistanceVariable(mesh, value = (-.5, .5, 1.5)) >>> ## answer = numerix.zeros((2, mesh.numberOfFaces),'d') >>> answer = FaceVariable(mesh=mesh, rank=1, value=0.).globalValue >>> answer[0, 7] = -1 >>> print(numerix.allclose(SurfactantConvectionVariable(distanceVar).globalValue, answer)) True
Change the dimensions:
>>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .25) >>> distanceVar = DistanceVariable(mesh, value = (-.25, .25, .75)) >>> answer[0, 7] = -.5 >>> print(numerix.allclose(SurfactantConvectionVariable(distanceVar).globalValue, answer)) True
Two dimensional example:
>>> mesh = Grid2D(nx = 2, ny = 2, dx = 1., dy = 1.) >>> distanceVar = DistanceVariable(mesh, value = (-1.5, -.5, -.5, .5)) >>> answer = FaceVariable(mesh=mesh, rank=1, value=0.).globalValue >>> answer[1, 2] = -.5 >>> answer[1, 3] = -1 >>> answer[0, 7] = -.5 >>> answer[0, 10] = -1 >>> print(numerix.allclose(SurfactantConvectionVariable(distanceVar).globalValue, answer)) True
Larger grid:
>>> mesh = Grid2D(nx = 3, ny = 3, dx = 1., dy = 1.) >>> distanceVar = DistanceVariable(mesh, value = (1.5, .5, 1.5, ... .5, -.5, .5, ... 1.5, .5, 1.5)) >>> answer = FaceVariable(mesh=mesh, rank=1, value=0.).globalValue >>> answer[1, 4] = .25 >>> answer[1, 7] = -.25 >>> answer[0, 17] = .25 >>> answer[0, 18] = -.25 >>> print(numerix.allclose(SurfactantConvectionVariable(distanceVar).globalValue, answer)) True
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__()¶
“Evaluate” the Variable and return its value
>>> a = Variable(value=3) >>> print(a()) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b() 7
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe _MeshVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new Variable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- cacheMe(recursive=False)¶
- constrain(value, where=None)¶
Constrain the Variable to have a value at an index or mask location specified by where.
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> print(v) [2 1 2 3] >>> v[:] = 10 >>> print(v) [ 2 10 10 10] >>> v.constrain(5, numerix.array((False, False, True, False))) >>> print(v) [ 2 10 5 10] >>> v[:] = 6 >>> print(v) [2 6 5 6] >>> v.constrain(8) >>> print(v) [8 8 8 8] >>> v[:] = 10 >>> print(v) [8 8 8 8] >>> del v.constraints[2] >>> print(v) [ 2 10 5 10]
>>> from fipy.variables.cellVariable import CellVariable >>> from fipy.meshes import Grid2D >>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v = CellVariable(mesh=m, rank=1, value=(x, y)) >>> v.constrain(((0.,), (-1.,)), where=m.facesLeft) >>> print(v.faceValue) [[ 0.5 1.5 0.5 1.5 0.5 1.5 0. 1. 1.5 0. 1. 1.5] [ 0.5 0.5 1. 1. 1.5 1.5 -1. 0.5 0.5 -1. 1.5 1.5]]
- Parameters:
value (float or array_like) – The value of the constraint
where (array_like of
bool) – The constraint mask or index specifying the location of the constraint
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- property divergence¶
the divergence of self,
,
- Returns:
divergence – one rank lower than self
- Return type:
Examples
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=3, ny=2) >>> from builtins import range >>> var = CellVariable(mesh=mesh, value=list(range(3*2))) >>> print(var.faceGrad.divergence) [ 4. 3. 2. -2. -3. -4.]
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property name¶
- property numericValue¶
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> v[:] = 10 >>> from fipy.boundaryConditions.constraint import Constraint >>> c1 = Constraint(5, numerix.array((False, False, True, False))) >>> v.constrain(c1) >>> v[:] = 6 >>> v.constrain(8) >>> v[:] = 10 >>> del v.constraints[2] >>> v.release(constraint=c1) >>> print(v) [ 2 10 10 10]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.surfactantVariable module¶
- class fipy.variables.surfactantVariable.SurfactantVariable(*args, **kwds)¶
Bases:
CellVariableThe SurfactantVariable maintains a conserved volumetric concentration on cells adjacent to, but in front of, the interface. The value argument corresponds to the initial concentration of surfactant on the interface (moles divided by area). The value held by the SurfactantVariable is actually a volume density (moles divided by volume).
A simple 1D test:
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(dx = 1., nx = 4) >>> from fipy.variables.distanceVariable import DistanceVariable >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (-1.5, -0.5, 0.5, 941.5)) >>> surfactantVariable = SurfactantVariable(value = 1, ... distanceVar = distanceVariable) >>> print(numerix.allclose(surfactantVariable, (0, 0., 1., 0))) 1
A 2D test case:
>>> from fipy.meshes import Grid2D >>> mesh = Grid2D(dx = 1., dy = 1., nx = 3, ny = 3) >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (1.5, 0.5, 1.5, ... 0.5, -0.5, 0.5, ... 1.5, 0.5, 1.5)) >>> surfactantVariable = SurfactantVariable(value = 1, ... distanceVar = distanceVariable) >>> print(numerix.allclose(surfactantVariable, (0, 1, 0, 1, 0, 1, 0, 1, 0))) 1
Another 2D test case:
>>> mesh = Grid2D(dx = .5, dy = .5, nx = 2, ny = 2) >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (-0.5, 0.5, 0.5, 1.5)) >>> surfactantVariable = SurfactantVariable(value = 1, ... distanceVar = distanceVariable) >>> print(numerix.allclose(surfactantVariable, ... (0, numerix.sqrt(2), numerix.sqrt(2), 0))) 1
- Parameters:
value (float or array_like) – The initial value.
distanceVar (DistanceVariable) –
name (str) – The name of the variable.
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- property interfaceVar¶
Returns the SurfactantVariable rendered as an _InterfaceSurfactantVariable which evaluates the surfactant concentration as an area concentration the interface rather than a volumetric concentration.
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.test module¶
Test numeric implementation of the mesh
fipy.variables.unaryOperatorVariable module¶
fipy.variables.uniformNoiseVariable module¶
- class fipy.variables.uniformNoiseVariable.UniformNoiseVariable(*args, **kwds)¶
Bases:
NoiseVariableRepresents a uniform distribution of random numbers.
We generate noise on a uniform Cartesian mesh
>>> from fipy.meshes import Grid2D >>> noise = UniformNoiseVariable(mesh=Grid2D(nx=100, ny=100))
and histogram the noise
>>> from fipy.variables.histogramVariable import HistogramVariable >>> histogram = HistogramVariable(distribution=noise, dx=0.01, nx=120, offset=-.1)
>>> if __name__ == '__main__': ... from fipy import Viewer ... viewer = Viewer(vars=noise, ... datamin=0, datamax=1) ... histoplot = Viewer(vars=histogram)
>>> from builtins import range >>> for i in range(10): ... noise.scramble() ... if __name__ == '__main__': ... viewer.plot() ... histoplot.plot()

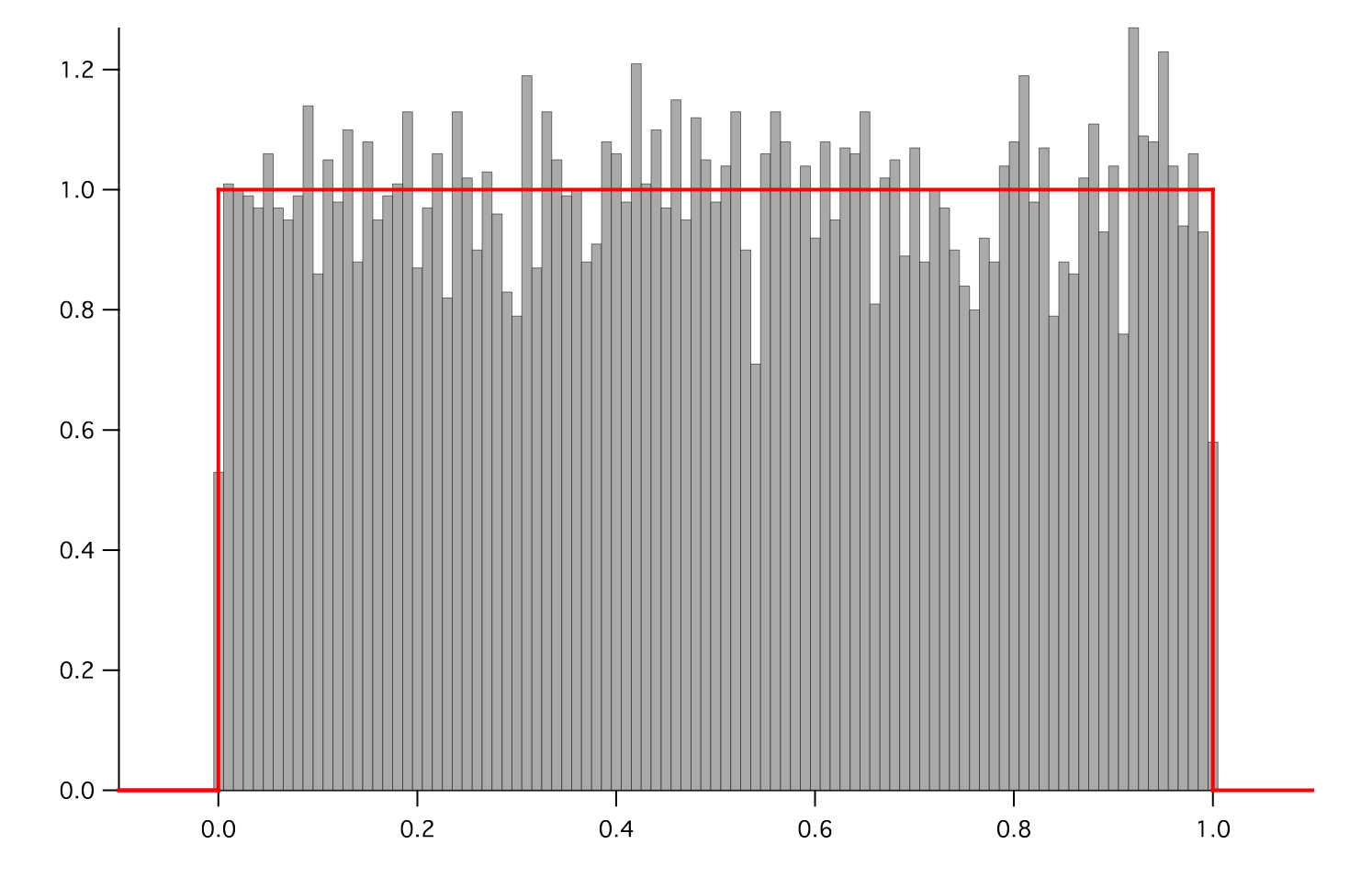
- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Copy the value of the NoiseVariable to a static CellVariable.
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- parallelRandom()¶
- put(indices, value)¶
- random()¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- scramble()¶
Generate a new random distribution.
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
fipy.variables.variable module¶
- class fipy.variables.variable.Variable(*args, **kwds)¶
Bases:
objectLazily evaluated quantity with units.
Using a
Variablein a mathematical expression will create an automatic dependencyVariable, e.g.,>>> a = Variable(value=3) >>> b = 4 * a >>> b (Variable(value=array(3)) * 4) >>> b() 12
Changes to the value of a
Variablewill automatically trigger changes in any dependentVariableobjects>>> a.setValue(5) >>> b (Variable(value=array(5)) * 4) >>> print(b()) 20
Create a Variable.
>>> Variable(value=3) Variable(value=array(3)) >>> Variable(value=3, unit="m") Variable(value=PhysicalField(3,'m')) >>> Variable(value=3, unit="m", array=numerix.zeros((3, 2), 'l')) Variable(value=PhysicalField(array([[3, 3], [3, 3], [3, 3]]),'m'))
- Parameters:
value (int or float or array_like) –
unit (str or PhysicalUnit) – The physical units of the variable
array (ndarray, optional) – The storage array for the Variable
name (str) – The user-readable name of the Variable
cached (bool) – whether to cache or always recalculate the value
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__()¶
“Evaluate” the Variable and return its value
>>> a = Variable(value=3) >>> print(a()) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b() 7
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe Variable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new Variable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- cacheMe(recursive=False)¶
- constrain(value, where=None)¶
Constrain the Variable to have a value at an index or mask location specified by where.
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> print(v) [2 1 2 3] >>> v[:] = 10 >>> print(v) [ 2 10 10 10] >>> v.constrain(5, numerix.array((False, False, True, False))) >>> print(v) [ 2 10 5 10] >>> v[:] = 6 >>> print(v) [2 6 5 6] >>> v.constrain(8) >>> print(v) [8 8 8 8] >>> v[:] = 10 >>> print(v) [8 8 8 8] >>> del v.constraints[2] >>> print(v) [ 2 10 5 10]
>>> from fipy.variables.cellVariable import CellVariable >>> from fipy.meshes import Grid2D >>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v = CellVariable(mesh=m, rank=1, value=(x, y)) >>> v.constrain(((0.,), (-1.,)), where=m.facesLeft) >>> print(v.faceValue) [[ 0.5 1.5 0.5 1.5 0.5 1.5 0. 1. 1.5 0. 1. 1.5] [ 0.5 0.5 1. 1. 1.5 1.5 -1. 0.5 0.5 -1. 1.5 1.5]]
- Parameters:
value (float or array_like) – The value of the constraint
where (array_like of
bool) – The constraint mask or index specifying the location of the constraint
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None, axis=0)¶
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- max(axis=None)¶
- min(axis=None)¶
- property name¶
- property numericValue¶
- put(indices, value)¶
- ravel()¶
- release(constraint)¶
Remove constraint from self
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> v[:] = 10 >>> from fipy.boundaryConditions.constraint import Constraint >>> c1 = Constraint(5, numerix.array((False, False, True, False))) >>> v.constrain(c1) >>> v[:] = 6 >>> v.constrain(8) >>> v[:] = 10 >>> del v.constraints[2] >>> v.release(constraint=c1) >>> print(v) [ 2 10 10 10]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
Tuple of array dimensions.
>>> Variable(value=3).shape () >>> numerix.allequal(Variable(value=(3,)).shape, (1,)) True >>> numerix.allequal(Variable(value=(3, 4)).shape, (2,)) True
>>> Variable(value="3 m").shape () >>> numerix.allequal(Variable(value=(3,), unit="m").shape, (1,)) True >>> numerix.allequal(Variable(value=(3, 4), unit="m").shape, (2,)) True
- std(axis=None, **kwargs)¶
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
Module contents¶
- class fipy.variables.BetaNoiseVariable(*args, **kwds)¶
Bases:
NoiseVariableRepresents a beta distribution of random numbers with the probability distribution
with a shape parameter
, a rate parameter
, and
.
Seed the random module for the sake of deterministic test results.
>>> from fipy import numerix >>> numerix.random.seed(1)
We generate noise on a uniform Cartesian mesh
>>> from fipy.variables.variable import Variable >>> alpha = Variable() >>> beta = Variable() >>> from fipy.meshes import Grid2D >>> noise = BetaNoiseVariable(mesh = Grid2D(nx = 100, ny = 100), alpha = alpha, beta = beta)
We histogram the root-volume-weighted noise distribution
>>> from fipy.variables.histogramVariable import HistogramVariable >>> histogram = HistogramVariable(distribution = noise, dx = 0.01, nx = 100)
and compare to a Gaussian distribution
>>> from fipy.variables.cellVariable import CellVariable >>> betadist = CellVariable(mesh = histogram.mesh) >>> x = CellVariable(mesh=histogram.mesh, value=histogram.mesh.cellCenters[0]) >>> from scipy.special import gamma as Gamma >>> betadist = ((Gamma(alpha + beta) / (Gamma(alpha) * Gamma(beta))) ... * x**(alpha - 1) * (1 - x)**(beta - 1))
>>> if __name__ == '__main__': ... from fipy import Viewer ... viewer = Viewer(vars=noise, datamin=0, datamax=1) ... histoplot = Viewer(vars=(histogram, betadist), ... datamin=0, datamax=1.5)
>>> from fipy.tools.numerix import arange
>>> for a in arange(0.5, 5, 0.5): ... alpha.value = a ... for b in arange(0.5, 5, 0.5): ... beta.value = b ... if __name__ == '__main__': ... import sys ... print("alpha: %g, beta: %g" % (alpha, beta), file=sys.stderr) ... viewer.plot() ... histoplot.plot()
>>> print(abs(noise.faceGrad.divergence.cellVolumeAverage) < 5e-15) 1


- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Copy the value of the NoiseVariable to a static CellVariable.
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- parallelRandom()¶
- put(indices, value)¶
- random()¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- scramble()¶
Generate a new random distribution.
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
- class fipy.variables.CellVariable(*args, **kwds)¶
Bases:
_MeshVariableRepresents the field of values of a variable on a Mesh.
A CellVariable can be
pickledto persistent storage (disk) for later use:>>> from fipy.meshes import Grid2D >>> mesh = Grid2D(dx = 1., dy = 1., nx = 10, ny = 10)
>>> var = CellVariable(mesh = mesh, value = 1., hasOld = 1, name = 'test') >>> x, y = mesh.cellCenters >>> var.value = (x * y)
>>> from fipy.tools import dump >>> (f, filename) = dump.write(var, extension = '.gz') >>> unPickledVar = dump.read(filename, f)
>>> print(var.allclose(unPickledVar, atol = 1e-10, rtol = 1e-10)) 1
- Parameters:
mesh (Mesh) – the mesh that defines the geometry of this Variable
name (str) – the user-readable name of the Variable
value (float or array_like) – the initial value
rank (int) – the rank (number of dimensions) of each element of this Variable. Default: 0
elementshape (
tupleofint) – the shape of each element of this variable Default: rank * (mesh.dim,)unit (str or PhysicalUnit) – The physical units of the variable
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
- class fipy.variables.DistanceVariable(*args, **kwds)¶
Bases:
CellVariableA DistanceVariable object calculates
so it satisfies,
using the fast marching method with an initial condition defined by the zero level set. The solution can either be first or second order.
Here we will define a few test cases. Firstly a 1D test case
>>> from fipy.meshes import Grid1D >>> from fipy.tools import serialComm >>> mesh = Grid1D(dx = .5, nx = 8, communicator=serialComm) >>> from .distanceVariable import DistanceVariable >>> var = DistanceVariable(mesh = mesh, value = (-1., -1., -1., -1., 1., 1., 1., 1.)) >>> var.calcDistanceFunction() >>> answer = (-1.75, -1.25, -.75, -0.25, 0.25, 0.75, 1.25, 1.75) >>> print(var.allclose(answer)) 1
A 1D test case with very small dimensions.
>>> dx = 1e-10 >>> mesh = Grid1D(dx = dx, nx = 8, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., -1., -1., -1., 1., 1., 1., 1.)) >>> var.calcDistanceFunction() >>> answer = numerix.arange(8) * dx - 3.5 * dx >>> print(var.allclose(answer)) 1
A 2D test case to test _calcTrialValue for a pathological case.
>>> dx = 1. >>> dy = 2. >>> from fipy.meshes import Grid2D >>> mesh = Grid2D(dx = dx, dy = dy, nx = 2, ny = 3, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., 1., 1., 1., -1., 1.))
>>> var.calcDistanceFunction() >>> vbl = -dx * dy / numerix.sqrt(dx**2 + dy**2) / 2. >>> vbr = dx / 2 >>> vml = dy / 2. >>> crossProd = dx * dy >>> dsq = dx**2 + dy**2 >>> top = vbr * dx**2 + vml * dy**2 >>> sqrt = crossProd**2 *(dsq - (vbr - vml)**2) >>> sqrt = numerix.sqrt(max(sqrt, 0)) >>> vmr = (top + sqrt) / dsq >>> answer = (vbl, vbr, vml, vmr, vbl, vbr) >>> print(var.allclose(answer)) 1
The extendVariable method solves the following equation for a given extensionVariable.
using the fast marching method with an initial condition defined at the zero level set.
>>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(dx = 1., dy = 1., nx = 2, ny = 2, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., 1., 1., 1.)) >>> var.calcDistanceFunction() >>> extensionVar = CellVariable(mesh = mesh, value = (-1, .5, 2, -1)) >>> tmp = 1 / numerix.sqrt(2) >>> print(var.allclose((-tmp / 2, 0.5, 0.5, 0.5 + tmp))) 1 >>> var.extendVariable(extensionVar, order=1) >>> print(extensionVar.allclose((1.25, .5, 2, 1.25))) 1 >>> mesh = Grid2D(dx = 1., dy = 1., nx = 3, ny = 3, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., 1., 1., ... 1., 1., 1., ... 1., 1., 1.)) >>> var.calcDistanceFunction(order=1) >>> extensionVar = CellVariable(mesh = mesh, value = (-1., .5, -1., ... 2., -1., -1., ... -1., -1., -1.))
>>> v1 = 0.5 + tmp >>> v2 = 1.5 >>> tmp1 = (v1 + v2) / 2 + numerix.sqrt(2. - (v1 - v2)**2) / 2 >>> tmp2 = tmp1 + 1 / numerix.sqrt(2) >>> print(var.allclose((-tmp / 2, 0.5, 1.5, 0.5, 0.5 + tmp, ... tmp1, 1.5, tmp1, tmp2))) 1 >>> answer = (1.25, .5, .5, 2, 1.25, 0.9544, 2, 1.5456, 1.25) >>> var.extendVariable(extensionVar, order=1) >>> print(extensionVar.allclose(answer, rtol = 1e-4)) 1
Test case for a bug that occurs when initializing the distance variable at the interface. Currently it is assumed that adjacent cells that are opposite sign neighbors have perpendicular normal vectors. In fact the two closest cells could have opposite normals.
>>> mesh = Grid1D(dx = 1., nx = 3, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., 1., -1.)) >>> var.calcDistanceFunction() >>> print(var.allclose((-0.5, 0.5, -0.5))) 1
Testing second order. This example failed with Scikit-fmm.
>>> mesh = Grid2D(dx = 1., dy = 1., nx = 4, ny = 4, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., -1., 1., 1., ... -1., -1., 1., 1., ... 1., 1., 1., 1., ... 1, 1, 1, 1)) >>> var.calcDistanceFunction(order=2) >>> answer = [-1.30473785, -0.5, 0.5, 1.49923009, ... -0.5, -0.35355339, 0.5, 1.45118446, ... 0.5, 0.5, 0.97140452, 1.76215286, ... 1.49923009, 1.45118446, 1.76215286, 2.33721352] >>> print(numerix.allclose(var, answer, rtol=1e-9)) True
** A test for a bug in both LSMLIB and Scikit-fmm **
The following test gives different result depending on whether LSMLIB or Scikit-fmm is used. There is a deeper problem that is related to this issue. When a value becomes “known” after previously being a “trial” value it updates its neighbors’ values. In a second order scheme the neighbors one step away also need to be updated (if the in between cell is “known” and the far cell is a “trial” cell), but are not in either package. By luck (due to trial values having the same value), the values calculated in Scikit-fmm for the following example are correct although an example that didn’t work for Scikit-fmm could also be constructed.
>>> mesh = Grid2D(dx = 1., dy = 1., nx = 4, ny = 4, communicator=serialComm) >>> var = DistanceVariable(mesh = mesh, value = (-1., -1., -1., -1., ... 1., 1., -1., -1., ... 1., 1., -1., -1., ... 1., 1., -1., -1.)) >>> var.calcDistanceFunction(order=2) >>> var.calcDistanceFunction(order=2) >>> answer = [-0.5, -0.58578644, -1.08578644, -1.85136395, ... 0.5, 0.29289322, -0.58578644, -1.54389939, ... 1.30473785, 0.5, -0.5, -1.5, ... 1.49547948, 0.5, -0.5, -1.5]
The 3rd and 7th element are different for LSMLIB. This is because the 15th element is not “known” when the “trial” value for the 7th element is calculated. Scikit-fmm calculates the values in a slightly different order so gets a seemingly better answer, but this is just chance.
>>> print(numerix.allclose(var, answer, rtol=1e-9)) True
Creates a distanceVariable object.
- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- calcDistanceFunction(order=2)¶
Calculates the distanceVariable as a distance function.
- Parameters:
order ({1, 2}) – The order of accuracy for the distance function calculation
- property cellInterfaceAreas¶
Returns the length of the interface that crosses the cell
A simple 1D test:
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(dx = 1., nx = 4) >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (-1.5, -0.5, 0.5, 1.5)) >>> answer = CellVariable(mesh=mesh, value=(0, 0., 1., 0)) >>> print(numerix.allclose(distanceVariable.cellInterfaceAreas, ... answer)) True
A 2D test case:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(dx = 1., dy = 1., nx = 3, ny = 3) >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (1.5, 0.5, 1.5, ... 0.5, -0.5, 0.5, ... 1.5, 0.5, 1.5)) >>> answer = CellVariable(mesh=mesh, ... value=(0, 1, 0, 1, 0, 1, 0, 1, 0)) >>> print(numerix.allclose(distanceVariable.cellInterfaceAreas, answer)) True
Another 2D test case:
>>> mesh = Grid2D(dx = .5, dy = .5, nx = 2, ny = 2) >>> from fipy.variables.cellVariable import CellVariable >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (-0.5, 0.5, 0.5, 1.5)) >>> answer = CellVariable(mesh=mesh, ... value=(0, numerix.sqrt(2) / 4, numerix.sqrt(2) / 4, 0)) >>> print(numerix.allclose(distanceVariable.cellInterfaceAreas, ... answer)) True
Test to check that the circumference of a circle is, in fact,
.
>>> mesh = Grid2D(dx = 0.05, dy = 0.05, nx = 20, ny = 20) >>> r = 0.25 >>> x, y = mesh.cellCenters >>> rad = numerix.sqrt((x - .5)**2 + (y - .5)**2) - r >>> distanceVariable = DistanceVariable(mesh = mesh, value = rad) >>> print(numerix.allclose(distanceVariable.cellInterfaceAreas.sum(), 1.57984690073)) 1
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- extendVariable(extensionVariable, order=2)¶
Calculates the extension of extensionVariable from the zero level set.
- Parameters:
extensionVariable (CellVariable) – The variable to extend from the zero level set.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getLSMshape()¶
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
- class fipy.variables.ExponentialNoiseVariable(*args, **kwds)¶
Bases:
NoiseVariableRepresents an exponential distribution of random numbers with the probability distribution
with a mean parameter
.
Seed the random module for the sake of deterministic test results.
>>> from fipy import numerix >>> numerix.random.seed(1)
We generate noise on a uniform Cartesian mesh
>>> from fipy.variables.variable import Variable >>> mean = Variable() >>> from fipy.meshes import Grid2D >>> noise = ExponentialNoiseVariable(mesh = Grid2D(nx = 100, ny = 100), mean = mean)
We histogram the root-volume-weighted noise distribution
>>> from fipy.variables.histogramVariable import HistogramVariable >>> histogram = HistogramVariable(distribution = noise, dx = 0.1, nx = 100)
and compare to a Gaussian distribution
>>> from fipy.variables.cellVariable import CellVariable >>> expdist = CellVariable(mesh = histogram.mesh) >>> x = histogram.mesh.cellCenters[0]
>>> if __name__ == '__main__': ... from fipy import Viewer ... viewer = Viewer(vars=noise, datamin=0, datamax=5) ... histoplot = Viewer(vars=(histogram, expdist), ... datamin=0, datamax=1.5)
>>> from fipy.tools.numerix import arange, exp
>>> for mu in arange(0.5, 3, 0.5): ... mean.value = (mu) ... expdist.value = ((1/mean)*exp(-x/mean)) ... if __name__ == '__main__': ... import sys ... print("mean: %g" % mean, file=sys.stderr) ... viewer.plot() ... histoplot.plot()
>>> print(abs(noise.faceGrad.divergence.cellVolumeAverage) < 5e-15) 1


- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Copy the value of the NoiseVariable to a static CellVariable.
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- parallelRandom()¶
- put(indices, value)¶
- random()¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- scramble()¶
Generate a new random distribution.
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
- class fipy.variables.FaceVariable(*args, **kwds)¶
Bases:
_MeshVariable- Parameters:
mesh (Mesh) – the mesh that defines the geometry of this Variable
name (str) – the user-readable name of the Variable
value (float or array_like) – the initial value
rank (int) – the rank (number of dimensions) of each element of this Variable. Default: 0
elementshape (
tupleofint) – the shape of each element of this variable Default: rank * (mesh.dim,)unit (str or PhysicalUnit) – The physical units of the variable
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__()¶
“Evaluate” the Variable and return its value
>>> a = Variable(value=3) >>> print(a()) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b() 7
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe _MeshVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new Variable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- cacheMe(recursive=False)¶
- constrain(value, where=None)¶
Constrain the Variable to have a value at an index or mask location specified by where.
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> print(v) [2 1 2 3] >>> v[:] = 10 >>> print(v) [ 2 10 10 10] >>> v.constrain(5, numerix.array((False, False, True, False))) >>> print(v) [ 2 10 5 10] >>> v[:] = 6 >>> print(v) [2 6 5 6] >>> v.constrain(8) >>> print(v) [8 8 8 8] >>> v[:] = 10 >>> print(v) [8 8 8 8] >>> del v.constraints[2] >>> print(v) [ 2 10 5 10]
>>> from fipy.variables.cellVariable import CellVariable >>> from fipy.meshes import Grid2D >>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v = CellVariable(mesh=m, rank=1, value=(x, y)) >>> v.constrain(((0.,), (-1.,)), where=m.facesLeft) >>> print(v.faceValue) [[ 0.5 1.5 0.5 1.5 0.5 1.5 0. 1. 1.5 0. 1. 1.5] [ 0.5 0.5 1. 1. 1.5 1.5 -1. 0.5 0.5 -1. 1.5 1.5]]
- Parameters:
value (float or array_like) – The value of the constraint
where (array_like of
bool) – The constraint mask or index specifying the location of the constraint
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- property divergence¶
the divergence of self,
,
- Returns:
divergence – one rank lower than self
- Return type:
Examples
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=3, ny=2) >>> from builtins import range >>> var = CellVariable(mesh=mesh, value=list(range(3*2))) >>> print(var.faceGrad.divergence) [ 4. 3. 2. -2. -3. -4.]
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property name¶
- property numericValue¶
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> v[:] = 10 >>> from fipy.boundaryConditions.constraint import Constraint >>> c1 = Constraint(5, numerix.array((False, False, True, False))) >>> v.constrain(c1) >>> v[:] = 6 >>> v.constrain(8) >>> v[:] = 10 >>> del v.constraints[2] >>> v.release(constraint=c1) >>> print(v) [ 2 10 10 10]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
- class fipy.variables.GammaNoiseVariable(*args, **kwds)¶
Bases:
NoiseVariableRepresents a gamma distribution of random numbers with the probability distribution
with a shape parameter
, a rate parameter
, and
.
Seed the random module for the sake of deterministic test results.
>>> from fipy import numerix >>> numerix.random.seed(1)
We generate noise on a uniform Cartesian mesh
>>> from fipy.variables.variable import Variable >>> alpha = Variable() >>> beta = Variable() >>> from fipy.meshes import Grid2D >>> noise = GammaNoiseVariable(mesh = Grid2D(nx = 100, ny = 100), shape = alpha, rate = beta)
We histogram the root-volume-weighted noise distribution
>>> from fipy.variables.histogramVariable import HistogramVariable >>> histogram = HistogramVariable(distribution = noise, dx = 0.1, nx = 300)
and compare to a Gaussian distribution
>>> from fipy.variables.cellVariable import CellVariable >>> x = CellVariable(mesh=histogram.mesh, value=histogram.mesh.cellCenters[0]) >>> from scipy.special import gamma as Gamma >>> from fipy.tools.numerix import exp >>> gammadist = (x**(alpha - 1) * (beta**alpha * exp(-beta * x)) / Gamma(alpha))
>>> if __name__ == '__main__': ... from fipy import Viewer ... viewer = Viewer(vars=noise, datamin=0, datamax=30) ... histoplot = Viewer(vars=(histogram, gammadist), ... datamin=0, datamax=1)
>>> from fipy.tools.numerix import arange
>>> for shape in arange(1, 8, 1): ... alpha.value = shape ... for rate in arange(0.5, 2.5, 0.5): ... beta.value = rate ... if __name__ == '__main__': ... import sys ... print("alpha: %g, beta: %g" % (alpha, beta), file=sys.stderr) ... viewer.plot() ... histoplot.plot()
>>> print(abs(noise.faceGrad.divergence.cellVolumeAverage) < 5e-15) 1


- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Copy the value of the NoiseVariable to a static CellVariable.
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- parallelRandom()¶
- put(indices, value)¶
- random()¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- scramble()¶
Generate a new random distribution.
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
- class fipy.variables.GaussianNoiseVariable(*args, **kwds)¶
Bases:
NoiseVariableRepresents a normal (Gaussian) distribution of random numbers with mean
and variance
, which has the probability distribution
For example, the variance of thermal noise that is uncorrelated in space and time is often expressed as
which can be obtained with:
sigmaSqrd = Mobility * kBoltzmann * Temperature / (mesh.cellVolumes * timeStep) GaussianNoiseVariable(mesh = mesh, variance = sigmaSqrd)
Note
If the time step will change as the simulation progresses, either through use of an adaptive iterator or by making manual changes at different stages, remember to declare timeStep as a Variable and to change its value with its setValue() method.
>>> import sys >>> from fipy.tools.numerix import *
>>> mean = 0. >>> variance = 4.
Seed the random module for the sake of deterministic test results.
>>> from fipy import numerix >>> numerix.random.seed(3)
We generate noise on a non-uniform Cartesian mesh with cell dimensions of
and
.
>>> from fipy.meshes import Grid2D >>> mesh = Grid2D(dx = arange(0.1, 5., 0.1)**2, dy = arange(0.1, 3., 0.1)**3) >>> from fipy.variables.cellVariable import CellVariable >>> volumes = CellVariable(mesh=mesh, value=mesh.cellVolumes) >>> noise = GaussianNoiseVariable(mesh = mesh, mean = mean, ... variance = variance / volumes)
We histogram the root-volume-weighted noise distribution
>>> from fipy.variables.histogramVariable import HistogramVariable >>> histogram = HistogramVariable(distribution = noise * sqrt(volumes), ... dx = 0.1, nx = 600, offset = -30)
and compare to a Gaussian distribution
>>> gauss = CellVariable(mesh = histogram.mesh) >>> x = histogram.mesh.cellCenters[0] >>> gauss.value = ((1/(sqrt(variance * 2 * pi))) * exp(-(x - mean)**2 / (2 * variance)))
>>> if __name__ == '__main__': ... from fipy.viewers import Viewer ... viewer = Viewer(vars=noise, ... datamin=-5, datamax=5) ... histoplot = Viewer(vars=(histogram, gauss))
>>> from builtins import range >>> for i in range(10): ... noise.scramble() ... if __name__ == '__main__': ... viewer.plot() ... histoplot.plot()
>>> print(abs(noise.faceGrad.divergence.cellVolumeAverage) < 5e-15) 1
Note that the noise exhibits larger amplitude in the small cells than in the large ones

but that the root-volume-weighted histogram is Gaussian.

- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Copy the value of the NoiseVariable to a static CellVariable.
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- parallelRandom()¶
- put(indices, value)¶
- random()¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- scramble()¶
Generate a new random distribution.
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
- class fipy.variables.HistogramVariable(*args, **kwds)¶
Bases:
CellVariableProduces a histogram of the values of the supplied distribution.
- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
- class fipy.variables.ModularVariable(*args, **kwds)¶
Bases:
CellVariableThe ModularVariable defines a variable that exists on the circle between
and
The following examples show how ModularVariable works. When subtracting the answer wraps back around the circle.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.tools import numerix >>> pi = numerix.pi >>> v1 = ModularVariable(mesh = mesh, value = (2*pi/3, -2*pi/3)) >>> v2 = ModularVariable(mesh = mesh, value = -2*pi/3) >>> print(numerix.allclose(v2 - v1, (2*pi/3, 0))) 1
Obtaining the arithmetic face value.
>>> print(numerix.allclose(v1.arithmeticFaceValue, (2*pi/3, pi, -2*pi/3))) 1
Obtaining the gradient.
>>> print(numerix.allclose(v1.grad, ((pi/3, pi/3),))) 1
Obtaining the gradient at the faces.
>>> print(numerix.allclose(v1.faceGrad, ((0, 2*pi/3, 0),))) 1
Obtaining the gradient at the faces but without modular arithmetic.
>>> print(numerix.allclose(v1.faceGradNoMod, ((0, -4*pi/3, 0),))) 1
- Parameters:
mesh (Mesh) – the mesh that defines the geometry of this Variable
name (str) – the user-readable name of the Variable
value (float or array_like) – the initial value
rank (int) – the rank (number of dimensions) of each element of this Variable. Default: 0
elementshape (
tupleofint) – the shape of each element of this variable Default: rank * (mesh.dim,)unit (str or PhysicalUnit) – The physical units of the variable
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
Adjusted for a ModularVariable
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable (second-order gradient). Adjusted for a ModularVariable
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceGradNoMod¶
Return
as a rank-1 FaceVariable (second-order gradient). Not adjusted for a ModularVariable
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient). Adjusted for a ModularVariable
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values. Test case due to bug.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 1) >>> var = ModularVariable(mesh=mesh, value=1., hasOld=1) >>> var.updateOld() >>> var[:] = 2 >>> answer = CellVariable(mesh=mesh, value=1.) >>> print(var.old.allclose(answer)) True
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
- class fipy.variables.ScharfetterGummelFaceVariable(*args, **kwds)¶
Bases:
_CellToFaceVariable- Parameters:
mesh (Mesh) – the mesh that defines the geometry of this Variable
name (str) – the user-readable name of the Variable
value (float or array_like) – the initial value
rank (int) – the rank (number of dimensions) of each element of this Variable. Default: 0
elementshape (
tupleofint) – the shape of each element of this variable Default: rank * (mesh.dim,)unit (str or PhysicalUnit) – The physical units of the variable
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__()¶
“Evaluate” the Variable and return its value
>>> a = Variable(value=3) >>> print(a()) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b() 7
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe _MeshVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new Variable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- cacheMe(recursive=False)¶
- constrain(value, where=None)¶
Constrain the Variable to have a value at an index or mask location specified by where.
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> print(v) [2 1 2 3] >>> v[:] = 10 >>> print(v) [ 2 10 10 10] >>> v.constrain(5, numerix.array((False, False, True, False))) >>> print(v) [ 2 10 5 10] >>> v[:] = 6 >>> print(v) [2 6 5 6] >>> v.constrain(8) >>> print(v) [8 8 8 8] >>> v[:] = 10 >>> print(v) [8 8 8 8] >>> del v.constraints[2] >>> print(v) [ 2 10 5 10]
>>> from fipy.variables.cellVariable import CellVariable >>> from fipy.meshes import Grid2D >>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v = CellVariable(mesh=m, rank=1, value=(x, y)) >>> v.constrain(((0.,), (-1.,)), where=m.facesLeft) >>> print(v.faceValue) [[ 0.5 1.5 0.5 1.5 0.5 1.5 0. 1. 1.5 0. 1. 1.5] [ 0.5 0.5 1. 1. 1.5 1.5 -1. 0.5 0.5 -1. 1.5 1.5]]
- Parameters:
value (float or array_like) – The value of the constraint
where (array_like of
bool) – The constraint mask or index specifying the location of the constraint
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- property divergence¶
the divergence of self,
,
- Returns:
divergence – one rank lower than self
- Return type:
Examples
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=3, ny=2) >>> from builtins import range >>> var = CellVariable(mesh=mesh, value=list(range(3*2))) >>> print(var.faceGrad.divergence) [ 4. 3. 2. -2. -3. -4.]
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property name¶
- property numericValue¶
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c0 = Constraint(0., where=m.facesLeft) >>> v.constrain(c0) >>> c1 = Constraint(3., where=m.facesRight) >>> v.faceValue.constrain(c1) >>> print(v.faceValue) [ 0. 1. 2. 3.] >>> v.faceValue.release(constraint=c0) >>> print(v.faceValue) [ 0.5 1. 2. 3. ] >>> v.faceValue.release(constraint=c1) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
- class fipy.variables.SurfactantConvectionVariable(*args, **kwds)¶
Bases:
FaceVariableConvection coefficient for the ConservativeSurfactantEquation. The coefficient only has a value for a negative distanceVar.
Simple one dimensional test:
>>> from fipy.variables.cellVariable import CellVariable >>> from fipy.meshes import Grid2D >>> mesh = Grid2D(nx = 3, ny = 1, dx = 1., dy = 1.) >>> from fipy.variables.distanceVariable import DistanceVariable >>> distanceVar = DistanceVariable(mesh, value = (-.5, .5, 1.5)) >>> ## answer = numerix.zeros((2, mesh.numberOfFaces),'d') >>> answer = FaceVariable(mesh=mesh, rank=1, value=0.).globalValue >>> answer[0, 7] = -1 >>> print(numerix.allclose(SurfactantConvectionVariable(distanceVar).globalValue, answer)) True
Change the dimensions:
>>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .25) >>> distanceVar = DistanceVariable(mesh, value = (-.25, .25, .75)) >>> answer[0, 7] = -.5 >>> print(numerix.allclose(SurfactantConvectionVariable(distanceVar).globalValue, answer)) True
Two dimensional example:
>>> mesh = Grid2D(nx = 2, ny = 2, dx = 1., dy = 1.) >>> distanceVar = DistanceVariable(mesh, value = (-1.5, -.5, -.5, .5)) >>> answer = FaceVariable(mesh=mesh, rank=1, value=0.).globalValue >>> answer[1, 2] = -.5 >>> answer[1, 3] = -1 >>> answer[0, 7] = -.5 >>> answer[0, 10] = -1 >>> print(numerix.allclose(SurfactantConvectionVariable(distanceVar).globalValue, answer)) True
Larger grid:
>>> mesh = Grid2D(nx = 3, ny = 3, dx = 1., dy = 1.) >>> distanceVar = DistanceVariable(mesh, value = (1.5, .5, 1.5, ... .5, -.5, .5, ... 1.5, .5, 1.5)) >>> answer = FaceVariable(mesh=mesh, rank=1, value=0.).globalValue >>> answer[1, 4] = .25 >>> answer[1, 7] = -.25 >>> answer[0, 17] = .25 >>> answer[0, 18] = -.25 >>> print(numerix.allclose(SurfactantConvectionVariable(distanceVar).globalValue, answer)) True
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__()¶
“Evaluate” the Variable and return its value
>>> a = Variable(value=3) >>> print(a()) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b() 7
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe _MeshVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new Variable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- cacheMe(recursive=False)¶
- constrain(value, where=None)¶
Constrain the Variable to have a value at an index or mask location specified by where.
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> print(v) [2 1 2 3] >>> v[:] = 10 >>> print(v) [ 2 10 10 10] >>> v.constrain(5, numerix.array((False, False, True, False))) >>> print(v) [ 2 10 5 10] >>> v[:] = 6 >>> print(v) [2 6 5 6] >>> v.constrain(8) >>> print(v) [8 8 8 8] >>> v[:] = 10 >>> print(v) [8 8 8 8] >>> del v.constraints[2] >>> print(v) [ 2 10 5 10]
>>> from fipy.variables.cellVariable import CellVariable >>> from fipy.meshes import Grid2D >>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v = CellVariable(mesh=m, rank=1, value=(x, y)) >>> v.constrain(((0.,), (-1.,)), where=m.facesLeft) >>> print(v.faceValue) [[ 0.5 1.5 0.5 1.5 0.5 1.5 0. 1. 1.5 0. 1. 1.5] [ 0.5 0.5 1. 1. 1.5 1.5 -1. 0.5 0.5 -1. 1.5 1.5]]
- Parameters:
value (float or array_like) – The value of the constraint
where (array_like of
bool) – The constraint mask or index specifying the location of the constraint
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- property divergence¶
the divergence of self,
,
- Returns:
divergence – one rank lower than self
- Return type:
Examples
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=3, ny=2) >>> from builtins import range >>> var = CellVariable(mesh=mesh, value=list(range(3*2))) >>> print(var.faceGrad.divergence) [ 4. 3. 2. -2. -3. -4.]
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property name¶
- property numericValue¶
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> v[:] = 10 >>> from fipy.boundaryConditions.constraint import Constraint >>> c1 = Constraint(5, numerix.array((False, False, True, False))) >>> v.constrain(c1) >>> v[:] = 6 >>> v.constrain(8) >>> v[:] = 10 >>> del v.constraints[2] >>> v.release(constraint=c1) >>> print(v) [ 2 10 10 10]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
- class fipy.variables.SurfactantVariable(*args, **kwds)¶
Bases:
CellVariableThe SurfactantVariable maintains a conserved volumetric concentration on cells adjacent to, but in front of, the interface. The value argument corresponds to the initial concentration of surfactant on the interface (moles divided by area). The value held by the SurfactantVariable is actually a volume density (moles divided by volume).
A simple 1D test:
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(dx = 1., nx = 4) >>> from fipy.variables.distanceVariable import DistanceVariable >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (-1.5, -0.5, 0.5, 941.5)) >>> surfactantVariable = SurfactantVariable(value = 1, ... distanceVar = distanceVariable) >>> print(numerix.allclose(surfactantVariable, (0, 0., 1., 0))) 1
A 2D test case:
>>> from fipy.meshes import Grid2D >>> mesh = Grid2D(dx = 1., dy = 1., nx = 3, ny = 3) >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (1.5, 0.5, 1.5, ... 0.5, -0.5, 0.5, ... 1.5, 0.5, 1.5)) >>> surfactantVariable = SurfactantVariable(value = 1, ... distanceVar = distanceVariable) >>> print(numerix.allclose(surfactantVariable, (0, 1, 0, 1, 0, 1, 0, 1, 0))) 1
Another 2D test case:
>>> mesh = Grid2D(dx = .5, dy = .5, nx = 2, ny = 2) >>> distanceVariable = DistanceVariable(mesh = mesh, ... value = (-0.5, 0.5, 0.5, 1.5)) >>> surfactantVariable = SurfactantVariable(value = 1, ... distanceVar = distanceVariable) >>> print(numerix.allclose(surfactantVariable, ... (0, numerix.sqrt(2), numerix.sqrt(2), 0))) 1
- Parameters:
value (float or array_like) – The initial value.
distanceVar (DistanceVariable) –
name (str) – The name of the variable.
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- property interfaceVar¶
Returns the SurfactantVariable rendered as an _InterfaceSurfactantVariable which evaluates the surfactant concentration as an area concentration the interface rather than a volumetric concentration.
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- put(indices, value)¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
- class fipy.variables.UniformNoiseVariable(*args, **kwds)¶
Bases:
NoiseVariableRepresents a uniform distribution of random numbers.
We generate noise on a uniform Cartesian mesh
>>> from fipy.meshes import Grid2D >>> noise = UniformNoiseVariable(mesh=Grid2D(nx=100, ny=100))
and histogram the noise
>>> from fipy.variables.histogramVariable import HistogramVariable >>> histogram = HistogramVariable(distribution=noise, dx=0.01, nx=120, offset=-.1)
>>> if __name__ == '__main__': ... from fipy import Viewer ... viewer = Viewer(vars=noise, ... datamin=0, datamax=1) ... histoplot = Viewer(vars=histogram)
>>> from builtins import range >>> for i in range(10): ... noise.scramble() ... if __name__ == '__main__': ... viewer.plot() ... histoplot.plot()


- Parameters:
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__(points=None, order=0, nearestCellIDs=None)¶
Interpolates the CellVariable to a set of points using a method that has a memory requirement on the order of Ncells by Npoints in general, but uses only Ncells when the CellVariable’s mesh is a UniformGrid object.
Tests
>>> from fipy import * >>> m = Grid2D(nx=3, ny=2) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)))) [ 0.5 1.5 1.5] >>> print(v(((0., 1.1, 1.2), (0., 1., 1.)), order=1)) [ 0.25 1.1 1.2 ] >>> m0 = Grid2D(nx=2, ny=2, dx=1., dy=1.) >>> m1 = Grid2D(nx=4, ny=4, dx=.5, dy=.5) >>> x, y = m0.cellCenters >>> v0 = CellVariable(mesh=m0, value=x * y) >>> print(v0(m1.cellCenters.globalValue)) [ 0.25 0.25 0.75 0.75 0.25 0.25 0.75 0.75 0.75 0.75 2.25 2.25 0.75 0.75 2.25 2.25] >>> print(v0(m1.cellCenters.globalValue, order=1)) [ 0.125 0.25 0.5 0.625 0.25 0.375 0.875 1. 0.5 0.875 1.875 2.25 0.625 1. 2.25 2.625]
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe CellVariable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new CellVariable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- property arithmeticFaceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- cacheMe(recursive=False)¶
- property cellVolumeAverage¶
Return the cell-volume-weighted average of the CellVariable:
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx = 3, ny = 1, dx = .5, dy = .1) >>> var = CellVariable(value = (1, 2, 6), mesh = mesh) >>> print(var.cellVolumeAverage) 3.0
- constrain(value, where=None)¶
Constrains the CellVariable to value at a location specified by where.
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> v.constrain(0., where=m.facesLeft) >>> v.faceGrad.constrain([1.], where=m.facesRight) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5]
Changing the constraint changes the dependencies
>>> v.constrain(1., where=m.facesLeft) >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can be Variable
>>> c = Variable(0.) >>> v.constrain(c, where=m.facesLeft) >>> print(v.faceGrad) [[ 1. 1. 1. 1.]] >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> c.value = 1. >>> print(v.faceGrad) [[-1. 1. 1. 1.]] >>> print(v.faceValue) [ 1. 1. 2. 2.5]
Constraints can have a Variable mask.
>>> v = CellVariable(mesh=m) >>> mask = FaceVariable(mesh=m, value=m.facesLeft) >>> v.constrain(1., where=mask) >>> print(v.faceValue) [ 1. 0. 0. 0.] >>> mask[:] = mask | m.facesRight >>> print(v.faceValue) [ 1. 0. 0. 1.]
- property constraintMask¶
Test that constraintMask returns a Variable that updates itself whenever the constraints change.
>>> from fipy import *
>>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v0 = CellVariable(mesh=m) >>> v0.constrain(1., where=m.facesLeft) >>> print(v0.faceValue.constraintMask) [False False False False False False True False False True False False] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 0. 1. 0. 0.] >>> v0.constrain(3., where=m.facesRight) >>> print(v0.faceValue.constraintMask) [False False False False False False True False True True False True] >>> print(v0.faceValue) [ 0. 0. 0. 0. 0. 0. 1. 0. 3. 1. 0. 3.] >>> v1 = CellVariable(mesh=m) >>> v1.constrain(1., where=(x < 1) & (y < 1)) >>> print(v1.constraintMask) [ True False False False] >>> print(v1) [ 1. 0. 0. 0.] >>> v1.constrain(3., where=(x > 1) & (y > 1)) >>> print(v1.constraintMask) [ True False False True] >>> print(v1) [ 1. 0. 0. 3.]
- property constraints¶
- copy()¶
Copy the value of the NoiseVariable to a static CellVariable.
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- property faceGrad¶
Return
as a rank-1 FaceVariable using differencing for the normal direction(second-order gradient).
- property faceGradAverage¶
Deprecated since version 3.3: use
grad.arithmeticFaceValue()insteadReturn
as a rank-1 FaceVariable using averaging for the normal direction(second-order gradient)
- property faceValue¶
Returns a FaceVariable whose value corresponds to the arithmetic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (0.5 / 1.) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (1.0 / 3.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.arithmeticFaceValue[mesh.interiorFaces.value] >>> answer = (R - L) * (5.0 / 55.0) + L >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- property gaussGrad¶
Return
as a rank-1 CellVariable (first-order gradient).
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- property globalValue¶
Concatenate and return values from all processors
When running on a single processor, the result is identical to
value.
- property grad¶
Return
as a rank-1 CellVariable (first-order gradient).
- property harmonicFaceValue¶
Returns a FaceVariable whose value corresponds to the harmonic interpolation of the adjacent cells:
>>> from fipy.meshes import Grid1D >>> from fipy import numerix >>> mesh = Grid1D(dx = (1., 1.)) >>> L = 1 >>> R = 2 >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (0.5 / 1.) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (2., 4.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (1.0 / 3.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
>>> mesh = Grid1D(dx = (10., 100.)) >>> var = CellVariable(mesh = mesh, value = (L, R)) >>> faceValue = var.harmonicFaceValue[mesh.interiorFaces.value] >>> answer = L * R / ((R - L) * (5.0 / 55.0) + L) >>> print(numerix.allclose(faceValue, answer, atol = 1e-10, rtol = 1e-10)) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- property leastSquaresGrad¶
Return
, which is determined by solving for
in the following matrix equation,
The matrix equation is derived by minimizing the following least squares sum,
Tests
>>> from fipy import Grid2D >>> m = Grid2D(nx=2, ny=2, dx=0.1, dy=2.0) >>> print(numerix.allclose(CellVariable(mesh=m, value=(0, 1, 3, 6)).leastSquaresGrad.globalValue, \ ... [[8.0, 8.0, 24.0, 24.0], ... [1.2, 2.0, 1.2, 2.0]])) True
>>> from fipy import Grid1D >>> print(numerix.allclose(CellVariable(mesh=Grid1D(dx=(2.0, 1.0, 0.5)), ... value=(0, 1, 2)).leastSquaresGrad.globalValue, [[0.461538461538, 0.8, 1.2]])) True
- max(axis=None)¶
- min(axis=None)¶
>>> from fipy import Grid2D, CellVariable >>> mesh = Grid2D(nx=5, ny=5) >>> x, y = mesh.cellCenters >>> v = CellVariable(mesh=mesh, value=x*y) >>> print(v.min()) 0.25
- property minmodFaceValue¶
Returns a FaceVariable with a value that is the minimum of the absolute values of the adjacent cells. If the values are of opposite sign then the result is zero:
>>> from fipy import * >>> print(CellVariable(mesh=Grid1D(nx=2), value=(1, 2)).minmodFaceValue) [1 1 2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, -2)).minmodFaceValue) [-1 -1 -2] >>> print(CellVariable(mesh=Grid1D(nx=2), value=(-1, 2)).minmodFaceValue) [-1 0 2]
- property name¶
- property numericValue¶
- property old¶
Return the values of the CellVariable from the previous solution sweep.
Combinations of CellVariable’s should also return old values.
>>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx = 2) >>> from fipy.variables.cellVariable import CellVariable >>> var1 = CellVariable(mesh = mesh, value = (2, 3), hasOld = 1) >>> var2 = CellVariable(mesh = mesh, value = (3, 4)) >>> v = var1 * var2 >>> print(v) [ 6 12] >>> var1.value = ((3, 2)) >>> print(v) [9 8] >>> print(v.old) [ 6 12]
The following small test is to correct for a bug when the operator does not just use variables.
>>> v1 = var1 * 3 >>> print(v1) [9 6] >>> print(v1.old) [6 9]
- parallelRandom()¶
- put(indices, value)¶
- random()¶
- property rank¶
- ravel()¶
- rdot(other, opShape=None, operatorClass=None)¶
Return the mesh-element–by–mesh-element (cell-by-cell, face-by-face, etc.) scalar product
Both self and other can be of arbitrary rank, and other does not need to be a _MeshVariable.
- release(constraint)¶
Remove constraint from self
>>> from fipy import * >>> m = Grid1D(nx=3) >>> v = CellVariable(mesh=m, value=m.cellCenters[0]) >>> c = Constraint(0., where=m.facesLeft) >>> v.constrain(c) >>> print(v.faceValue) [ 0. 1. 2. 2.5] >>> v.release(constraint=c) >>> print(v.faceValue) [ 0.5 1. 2. 2.5]
- scramble()¶
Generate a new random distribution.
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
>>> from fipy.meshes import Grid2D >>> from fipy.variables.cellVariable import CellVariable >>> mesh = Grid2D(nx=2, ny=3) >>> var = CellVariable(mesh=mesh) >>> print(numerix.allequal(var.shape, (6,))) True >>> print(numerix.allequal(var.arithmeticFaceValue.shape, (17,))) True >>> print(numerix.allequal(var.grad.shape, (2, 6))) True >>> print(numerix.allequal(var.faceGrad.shape, (2, 17))) True
Have to account for zero length arrays
>>> from fipy import Grid1D >>> m = Grid1D(nx=0) >>> v = CellVariable(mesh=m, elementshape=(2,)) >>> numerix.allequal((v * 1).shape, (2, 0)) True
- std(axis=None, **kwargs)¶
Evaluate standard deviation of all the elements of a MeshVariable.
Adapted from http://mpitutorial.com/tutorials/mpi-reduce-and-allreduce/
>>> import fipy as fp >>> mesh = fp.Grid2D(nx=2, ny=2, dx=2., dy=5.) >>> var = fp.CellVariable(value=(1., 2., 3., 4.), mesh=mesh) >>> print((var.std()**2).allclose(1.25)) True
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- updateOld()¶
Set the values of the previous solution sweep to the current values.
>>> from fipy import * >>> v = CellVariable(mesh=Grid1D(), hasOld=False) >>> v.updateOld() Traceback (most recent call last): ... AssertionError: The updateOld method requires the CellVariable to have an old value. Set hasOld to True when instantiating the CellVariable.
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
- class fipy.variables.Variable(*args, **kwds)¶
Bases:
objectLazily evaluated quantity with units.
Using a
Variablein a mathematical expression will create an automatic dependencyVariable, e.g.,>>> a = Variable(value=3) >>> b = 4 * a >>> b (Variable(value=array(3)) * 4) >>> b() 12
Changes to the value of a
Variablewill automatically trigger changes in any dependentVariableobjects>>> a.setValue(5) >>> b (Variable(value=array(5)) * 4) >>> print(b()) 20
Create a Variable.
>>> Variable(value=3) Variable(value=array(3)) >>> Variable(value=3, unit="m") Variable(value=PhysicalField(3,'m')) >>> Variable(value=3, unit="m", array=numerix.zeros((3, 2), 'l')) Variable(value=PhysicalField(array([[3, 3], [3, 3], [3, 3]]),'m'))
- Parameters:
value (int or float or array_like) –
unit (str or PhysicalUnit) – The physical units of the variable
array (ndarray, optional) – The storage array for the Variable
name (str) – The user-readable name of the Variable
cached (bool) – whether to cache or always recalculate the value
- __abs__()¶
Following test it to fix a bug with C inline string using abs() instead of fabs()
>>> print(abs(Variable(2.3) - Variable(1.2))) 1.1
Check representation works with different versions of numpy
>>> print(repr(abs(Variable(2.3)))) numerix.fabs(Variable(value=array(2.3)))
- __add__(other)¶
- __and__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) & (b == 1), [False, True, False, False]).all()) True >>> print(a & b) [0 0 0 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) & (b == 1), [False, True, False, False])) True >>> print(a & b) [0 0 0 1]
- __array__(t=None)¶
Attempt to convert the Variable to a numerix array object
>>> v = Variable(value=[2, 3]) >>> print(numerix.array(v)) [2 3]
A dimensional Variable will convert to the numeric value in its base units
>>> v = Variable(value=[2, 3], unit="mm") >>> numerix.array(v) array([ 0.002, 0.003])
- __array_priority__ = 100.0¶
- __array_wrap__(arr, context=None)¶
Required to prevent numpy not calling the reverse binary operations. Both the following tests are examples ufuncs.
>>> print(type(numerix.array([1.0, 2.0]) * Variable([1.0, 2.0]))) <class 'fipy.variables.binaryOperatorVariable...binOp'>
>>> from scipy.special import gamma as Gamma >>> print(type(Gamma(Variable([1.0, 2.0])))) <class 'fipy.variables.unaryOperatorVariable...unOp'>
- __bool__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __call__()¶
“Evaluate” the Variable and return its value
>>> a = Variable(value=3) >>> print(a()) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b() 7
- __div__(other)¶
- __eq__(other)¶
Test if a Variable is equal to another quantity
>>> a = Variable(value=3) >>> b = (a == 4) >>> b (Variable(value=array(3)) == 4) >>> b() 0
- __float__()¶
- __ge__(other)¶
Test if a Variable is greater than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a >= 4) >>> b (Variable(value=array(3)) >= 4) >>> b() 0 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 1
- __getitem__(index)¶
“Evaluate” the Variable and return the specified element
>>> a = Variable(value=((3., 4.), (5., 6.)), unit="m") + "4 m" >>> print(a[1, 1]) 10.0 m
It is an error to slice a Variable whose value is not sliceable
>>> Variable(value=3)[2] Traceback (most recent call last): ... IndexError: 0-d arrays can't be indexed
- __getstate__()¶
Used internally to collect the necessary information to
picklethe Variable to persistent storage.
- __gt__(other)¶
Test if a Variable is greater than another quantity
>>> a = Variable(value=3) >>> b = (a > 4) >>> b (Variable(value=array(3)) > 4) >>> print(b()) 0 >>> a.value = 5 >>> print(b()) 1
- __hash__()¶
Return hash(self).
- __int__()¶
- __invert__()¶
Returns logical “not” of the Variable
>>> a = Variable(value=True) >>> print(~a) False
- __iter__()¶
- __le__(other)¶
Test if a Variable is less than or equal to another quantity
>>> a = Variable(value=3) >>> b = (a <= 4) >>> b (Variable(value=array(3)) <= 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 1 >>> a.value = 5 >>> print(b()) 0
- __len__()¶
- __lt__(other)¶
Test if a Variable is less than another quantity
>>> a = Variable(value=3) >>> b = (a < 4) >>> b (Variable(value=array(3)) < 4) >>> b() 1 >>> a.value = 4 >>> print(b()) 0 >>> print(1000000000000000000 * Variable(1) < 1.) 0 >>> print(1000 * Variable(1) < 1.) 0
Python automatically reverses the arguments when necessary
>>> 4 > Variable(value=3) (Variable(value=array(3)) < 4)
- __mod__(other)¶
- __mul__(other)¶
- __ne__(other)¶
Test if a Variable is not equal to another quantity
>>> a = Variable(value=3) >>> b = (a != 4) >>> b (Variable(value=array(3)) != 4) >>> b() 1
- __neg__()¶
- static __new__(cls, *args, **kwds)¶
- __nonzero__()¶
>>> print(bool(Variable(value=0))) 0 >>> print(bool(Variable(value=(0, 0, 1, 1)))) Traceback (most recent call last): ... ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
- __or__(other)¶
This test case has been added due to a weird bug that was appearing.
>>> a = Variable(value=(0, 0, 1, 1)) >>> b = Variable(value=(0, 1, 0, 1)) >>> print(numerix.equal((a == 0) | (b == 1), [True, True, False, True]).all()) True >>> print(a | b) [0 1 1 1] >>> from fipy.meshes import Grid1D >>> mesh = Grid1D(nx=4) >>> from fipy.variables.cellVariable import CellVariable >>> a = CellVariable(value=(0, 0, 1, 1), mesh=mesh) >>> b = CellVariable(value=(0, 1, 0, 1), mesh=mesh) >>> print(numerix.allequal((a == 0) | (b == 1), [True, True, False, True])) True >>> print(a | b) [0 1 1 1]
- __pos__()¶
- __pow__(other)¶
return self**other, or self raised to power other
>>> print(Variable(1, "mol/l")**3) 1.0 mol**3/l**3 >>> print((Variable(1, "mol/l")**3).unit) <PhysicalUnit mol**3/l**3>
- __radd__(other)¶
- __rdiv__(other)¶
- __repr__()¶
Return repr(self).
- __rmul__(other)¶
- __rpow__(other)¶
- __rsub__(other)¶
- __rtruediv__(other)¶
- __setitem__(index, value)¶
- __setstate__(dict)¶
Used internally to create a new Variable from
pickledpersistent storage.
- __str__()¶
Return str(self).
- __sub__(other)¶
- __truediv__(other)¶
- all(axis=None)¶
>>> print(Variable(value=(0, 0, 1, 1)).all()) 0 >>> print(Variable(value=(1, 1, 1, 1)).all()) 1
- allclose(other, rtol=1e-05, atol=1e-08)¶
>>> var = Variable((1, 1)) >>> print(var.allclose((1, 1))) 1 >>> print(var.allclose((1,))) 1
The following test is to check that the system does not run out of memory.
>>> from fipy.tools import numerix >>> var = Variable(numerix.ones(10000)) >>> print(var.allclose(numerix.zeros(10000, 'l'))) False
- allequal(other)¶
- any(axis=None)¶
>>> print(Variable(value=0).any()) 0 >>> print(Variable(value=(0, 0, 1, 1)).any()) 1
- cacheMe(recursive=False)¶
- constrain(value, where=None)¶
Constrain the Variable to have a value at an index or mask location specified by where.
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> print(v) [2 1 2 3] >>> v[:] = 10 >>> print(v) [ 2 10 10 10] >>> v.constrain(5, numerix.array((False, False, True, False))) >>> print(v) [ 2 10 5 10] >>> v[:] = 6 >>> print(v) [2 6 5 6] >>> v.constrain(8) >>> print(v) [8 8 8 8] >>> v[:] = 10 >>> print(v) [8 8 8 8] >>> del v.constraints[2] >>> print(v) [ 2 10 5 10]
>>> from fipy.variables.cellVariable import CellVariable >>> from fipy.meshes import Grid2D >>> m = Grid2D(nx=2, ny=2) >>> x, y = m.cellCenters >>> v = CellVariable(mesh=m, rank=1, value=(x, y)) >>> v.constrain(((0.,), (-1.,)), where=m.facesLeft) >>> print(v.faceValue) [[ 0.5 1.5 0.5 1.5 0.5 1.5 0. 1. 1.5 0. 1. 1.5] [ 0.5 0.5 1. 1. 1.5 1.5 -1. 0.5 0.5 -1. 1.5 1.5]]
- Parameters:
value (float or array_like) – The value of the constraint
where (array_like of
bool) – The constraint mask or index specifying the location of the constraint
- property constraints¶
- copy()¶
Make an duplicate of the Variable
>>> a = Variable(value=3) >>> b = a.copy() >>> b Variable(value=array(3))
The duplicate will not reflect changes made to the original
>>> a.setValue(5) >>> b Variable(value=array(3))
Check that this works for arrays.
>>> a = Variable(value=numerix.array((0, 1, 2))) >>> b = a.copy() >>> b Variable(value=array([0, 1, 2])) >>> a[1] = 3 >>> b Variable(value=array([0, 1, 2]))
- dontCacheMe(recursive=False)¶
- dot(other, opShape=None, operatorClass=None, axis=0)¶
- getsctype(default=None)¶
Returns the Numpy sctype of the underlying array.
>>> Variable(1).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1)) True >>> Variable(1.).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array(1.)) True >>> Variable((1, 1.)).getsctype() == numerix.NUMERIX.obj2sctype(numerix.array((1., 1.))) True
- inBaseUnits()¶
Return the value of the Variable with all units reduced to their base SI elements.
>>> e = Variable(value="2.7 Hartree*Nav") >>> print(e.inBaseUnits().allclose("7088849.01085 kg*m**2/s**2/mol")) 1
- inUnitsOf(*units)¶
Returns one or more Variable objects that express the same physical quantity in different units. The units are specified by strings containing their names. The units must be compatible with the unit of the object. If one unit is specified, the return value is a single Variable.
>>> freeze = Variable('0 degC') >>> print(freeze.inUnitsOf('degF').allclose("32.0 degF")) 1
If several units are specified, the return value is a tuple of Variable instances with with one element per unit such that the sum of all quantities in the tuple equals the the original quantity and all the values except for the last one are integers. This is used to convert to irregular unit systems like hour/minute/second. The original object will not be changed.
>>> t = Variable(value=314159., unit='s') >>> from builtins import zip >>> print(numerix.allclose([e.allclose(v) for (e, v) in zip(t.inUnitsOf('d', 'h', 'min', 's'), ... ['3.0 d', '15.0 h', '15.0 min', '59.0 s'])], ... True)) 1
- itemset(value)¶
- property itemsize¶
- max(axis=None)¶
- min(axis=None)¶
- property name¶
- property numericValue¶
- put(indices, value)¶
- ravel()¶
- release(constraint)¶
Remove constraint from self
>>> v = Variable((0, 1, 2, 3)) >>> v.constrain(2, numerix.array((True, False, False, False))) >>> v[:] = 10 >>> from fipy.boundaryConditions.constraint import Constraint >>> c1 = Constraint(5, numerix.array((False, False, True, False))) >>> v.constrain(c1) >>> v[:] = 6 >>> v.constrain(8) >>> v[:] = 10 >>> del v.constraints[2] >>> v.release(constraint=c1) >>> print(v) [ 2 10 10 10]
- setValue(value, unit=None, where=None)¶
Set the value of the Variable. Can take a masked array.
>>> a = Variable((1, 2, 3)) >>> a.setValue(5, where=(1, 0, 1)) >>> print(a) [5 2 5]
>>> b = Variable((4, 5, 6)) >>> a.setValue(b, where=(1, 0, 1)) >>> print(a) [4 2 6] >>> print(b) [4 5 6] >>> a.value = 3 >>> print(a) [3 3 3]
>>> b = numerix.array((3, 4, 5)) >>> a.value = b >>> a[:] = 1 >>> print(b) [3 4 5]
>>> a.setValue((4, 5, 6), where=(1, 0)) Traceback (most recent call last): .... ValueError: shape mismatch: objects cannot be broadcast to a single shape
- property shape¶
Tuple of array dimensions.
>>> Variable(value=3).shape () >>> numerix.allequal(Variable(value=(3,)).shape, (1,)) True >>> numerix.allequal(Variable(value=(3, 4)).shape, (2,)) True
>>> Variable(value="3 m").shape () >>> numerix.allequal(Variable(value=(3,), unit="m").shape, (1,)) True >>> numerix.allequal(Variable(value=(3, 4), unit="m").shape, (2,)) True
- std(axis=None, **kwargs)¶
- property subscribedVariables¶
- sum(axis=None)¶
- take(ids, axis=0)¶
- tostring(max_line_width=75, precision=8, suppress_small=False, separator=' ')¶
- property unit¶
Return the unit object of self.
>>> Variable(value="1 m").unit <PhysicalUnit m>
- property value¶
“Evaluate” the Variable and return its value (longhand)
>>> a = Variable(value=3) >>> print(a.value) 3 >>> b = a + 4 >>> b (Variable(value=array(3)) + 4) >>> b.value 7
 FiPy
FiPy