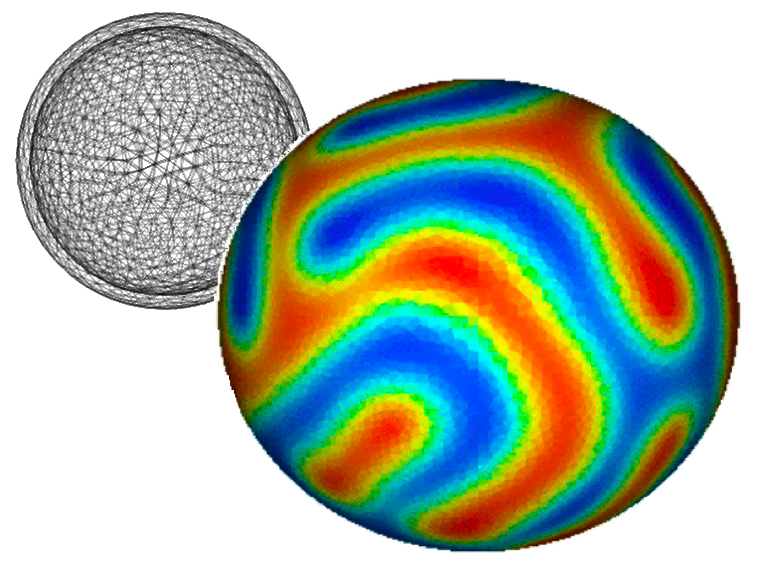
examples.cahnHilliard.sphere¶
Solves the Cahn-Hilliard problem on the surface of a sphere.
This phenomenon can occur on vesicles (http://www.youtube.com/watch?v=kDsFP67_ZSE).
>>> from fipy import CellVariable, Gmsh2DIn3DSpace, GaussianNoiseVariable, Viewer, TransientTerm, DiffusionTerm, DefaultSolver
>>> from fipy.tools import numerix
The only difference from examples.cahnHilliard.mesh2D is the
declaration of mesh.
>>> mesh = Gmsh2DIn3DSpace('''
... radius = 5.0;
... cellSize = 0.3;
...
... // create inner 1/8 shell
... Point(1) = {0, 0, 0, cellSize};
... Point(2) = {-radius, 0, 0, cellSize};
... Point(3) = {0, radius, 0, cellSize};
... Point(4) = {0, 0, radius, cellSize};
... Circle(1) = {2, 1, 3};
... Circle(2) = {4, 1, 2};
... Circle(3) = {4, 1, 3};
... Line Loop(1) = {1, -3, 2} ;
... Ruled Surface(1) = {1};
...
... // create remaining 7/8 inner shells
... t1[] = Rotate {{0,0,1},{0,0,0},Pi/2} {Duplicata{Surface{1};}};
... t2[] = Rotate {{0,0,1},{0,0,0},Pi} {Duplicata{Surface{1};}};
... t3[] = Rotate {{0,0,1},{0,0,0},Pi*3/2} {Duplicata{Surface{1};}};
... t4[] = Rotate {{0,1,0},{0,0,0},-Pi/2} {Duplicata{Surface{1};}};
... t5[] = Rotate {{0,0,1},{0,0,0},Pi/2} {Duplicata{Surface{t4[0]};}};
... t6[] = Rotate {{0,0,1},{0,0,0},Pi} {Duplicata{Surface{t4[0]};}};
... t7[] = Rotate {{0,0,1},{0,0,0},Pi*3/2} {Duplicata{Surface{t4[0]};}};
...
... // create entire inner and outer shell
... Surface Loop(100)={1,t1[0],t2[0],t3[0],t7[0],t4[0],t5[0],t6[0]};
... ''', overlap=2).extrude(extrudeFunc=lambda r: 1.1 * r)
>>> phi = CellVariable(name=r"$\phi$", mesh=mesh)
We start the problem with random fluctuations about
>>> phi.setValue(GaussianNoiseVariable(mesh=mesh,
... mean=0.5,
... variance=0.01))
FiPy doesn’t plot or output anything unless you tell it to: If
MayaviClient is available, we
can customize the view with a subclass of
MayaviDaemon.
>>> if __name__ == "__main__":
... try:
... viewer = MayaviClient(vars=phi,
... datamin=0., datamax=1.,
... daemon_file="examples/cahnHilliard/sphereDaemon.py")
... except:
... viewer = Viewer(vars=phi,
... datamin=0., datamax=1.,
... xmin=-2.5, zmax=2.5)
For FiPy, we need to perform the partial derivative
manually and then put the equation in the canonical
form by decomposing the spatial derivatives
so that each
Term is of a single, even order:
FiPy would automatically interpolate
D * a**2 * (1 - 6 * phi * (1 - phi))
onto the faces, where the diffusive flux is calculated, but we obtain
somewhat more accurate results by performing a linear interpolation from
phi at cell centers to PHI at face centers.
Some problems benefit from non-linear interpolations, such as harmonic or
geometric means, and FiPy makes it easy to obtain these, too.
>>> PHI = phi.arithmeticFaceValue
>>> D = a = epsilon = 1.
>>> eq = (TransientTerm()
... == DiffusionTerm(coeff=D * a**2 * (1 - 6 * PHI * (1 - PHI)))
... - DiffusionTerm(coeff=(D, epsilon**2)))
Because the evolution of a spinodal microstructure slows with time, we use exponentially increasing time steps to keep the simulation “interesting”. The FiPy user always has direct control over the evolution of their problem.
>>> dexp = -5
>>> elapsed = 0.
>>> if __name__ == "__main__":
... duration = 1000.
... else:
... duration = 1e-2
>>> while elapsed < duration:
... dt = min(100, numerix.exp(dexp))
... elapsed += dt
... dexp += 0.01
... eq.solve(phi, dt=dt, solver=DefaultSolver(precon=None))
... if __name__ == "__main__":
... viewer.plot()
 FiPy
FiPy